You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004365_01080
You are here: Home > Sequence: MGYG000004365_01080
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000004365_01080 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13240; End: 15399 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 457 | 713 | 3.8e-48 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 5.03e-38 | 457 | 711 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 9.82e-37 | 457 | 713 | 96 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.09e-22 | 455 | 717 | 117 | 343 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 2.07e-10 | 54 | 151 | 13 | 103 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 4.34e-06 | 76 | 153 | 58 | 129 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANU56191.1 | 1.21e-252 | 1 | 715 | 10 | 743 |
| QQR18969.1 | 1.21e-252 | 1 | 715 | 10 | 743 |
| QRX63290.1 | 1.78e-250 | 7 | 717 | 19 | 731 |
| ASB48786.1 | 1.18e-236 | 11 | 719 | 21 | 736 |
| QCD34661.1 | 1.70e-233 | 1 | 718 | 11 | 766 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CLX_A | 1.21e-18 | 464 | 718 | 103 | 347 | CatalyticCore Of Xylanase A [Cellvibrio japonicus],1CLX_B Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_C Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_D Catalytic Core Of Xylanase A [Cellvibrio japonicus] |
| 1W2P_A | 1.22e-18 | 464 | 718 | 104 | 348 | The3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2P_B The 3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W2V_A | 1.22e-18 | 464 | 718 | 104 | 348 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2V_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W32_A | 1.22e-18 | 464 | 718 | 104 | 348 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W3H_A | 1.40e-18 | 464 | 718 | 115 | 359 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W3H_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P14768 | 3.42e-17 | 464 | 718 | 367 | 611 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| P23360 | 5.96e-16 | 461 | 715 | 130 | 327 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| Q9HEZ1 | 9.69e-16 | 477 | 715 | 211 | 406 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
| B7SIW2 | 9.08e-15 | 477 | 716 | 201 | 398 | Endo-1,4-beta-xylanase C OS=Phanerodontia chrysosporium OX=2822231 GN=xynC PE=1 SV=1 |
| P10478 | 2.07e-14 | 465 | 717 | 622 | 836 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
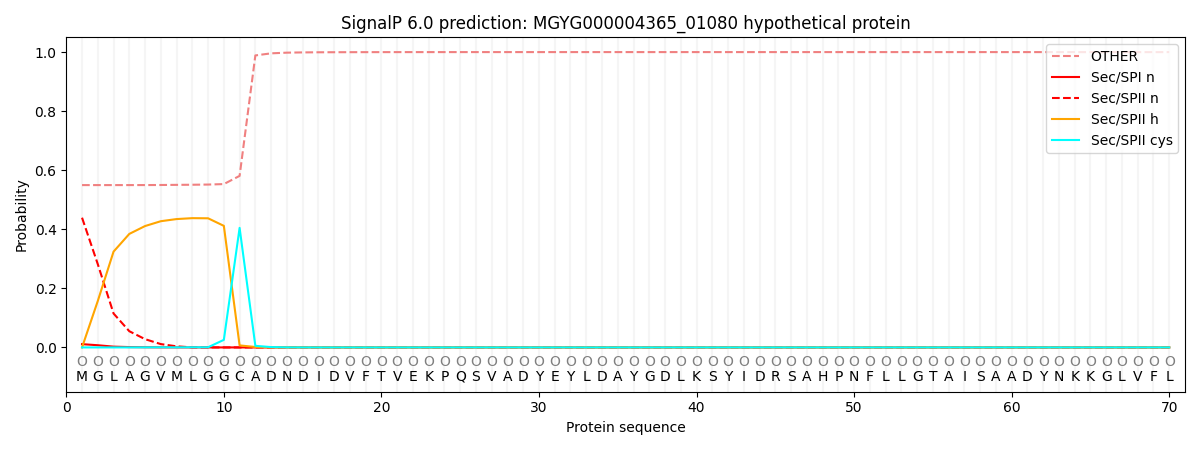
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.550513 | 0.010128 | 0.439354 | 0.000017 | 0.000014 | 0.000014 |
