You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004365_01084
You are here: Home > Sequence: MGYG000004365_01084
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000004365_01084 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19347; End: 21416 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 48 | 325 | 1.5e-113 | 0.9884169884169884 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.08e-12 | 42 | 276 | 2 | 201 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 0.004 | 1 | 207 | 6 | 200 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| TIGR04183 | Por_Secre_tail | 0.005 | 633 | 688 | 6 | 69 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD43724.1 | 0.0 | 24 | 685 | 10 | 666 |
| AHW58816.1 | 4.37e-238 | 1 | 685 | 1 | 675 |
| ADD61482.1 | 4.30e-226 | 30 | 625 | 30 | 630 |
| ADD61488.1 | 4.30e-226 | 30 | 625 | 30 | 630 |
| QRM99007.1 | 8.28e-226 | 30 | 625 | 50 | 649 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P06564 | 6.21e-07 | 35 | 272 | 62 | 266 | Endoglucanase OS=Alkalihalobacillus akibai (strain ATCC 43226 / DSM 21942 / CIP 109018 / JCM 9157 / 1139) OX=1236973 PE=1 SV=1 |
| P19570 | 6.28e-07 | 33 | 459 | 79 | 463 | Endoglucanase C OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
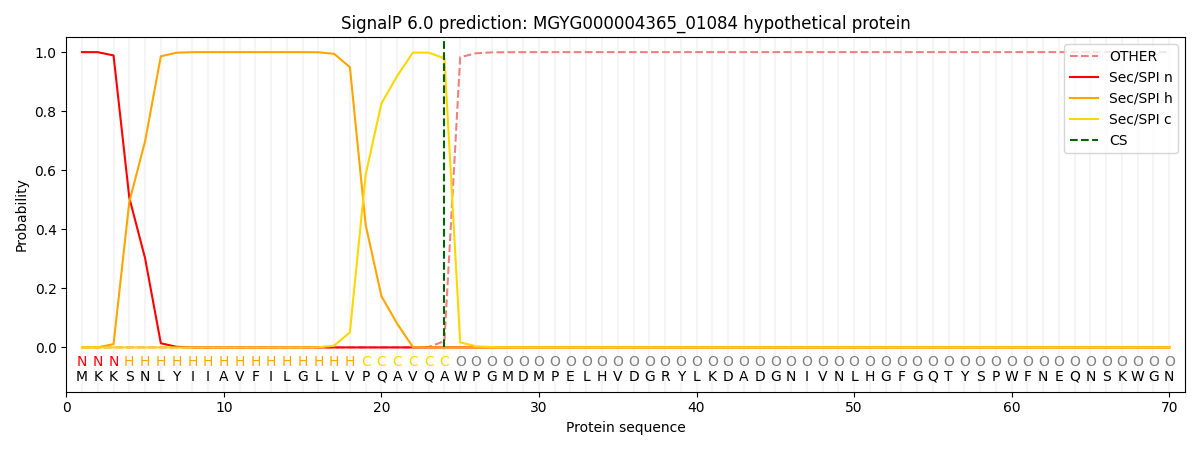
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000211 | 0.999111 | 0.000173 | 0.000167 | 0.000159 | 0.000146 |

