You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004369_00615
You are here: Home > Sequence: MGYG000004369_00615
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-882 sp000435595 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-882; CAG-882 sp000435595 | |||||||||||
| CAZyme ID | MGYG000004369_00615 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 70373; End: 73174 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 78 | 350 | 1.7e-88 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.20e-53 | 79 | 350 | 16 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.49e-24 | 53 | 353 | 45 | 363 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG2723 | BglB | 4.31e-05 | 99 | 197 | 72 | 168 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| COG5492 | YjdB | 1.83e-04 | 386 | 561 | 161 | 328 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 5.19e-04 | 509 | 565 | 25 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18792.1 | 4.42e-210 | 40 | 820 | 243 | 1037 |
| AUO19859.1 | 3.27e-114 | 47 | 557 | 109 | 633 |
| BCN29385.1 | 7.84e-76 | 47 | 730 | 349 | 950 |
| CCO05502.1 | 9.36e-74 | 1 | 384 | 1 | 399 |
| ADX05698.1 | 1.45e-70 | 49 | 385 | 9 | 378 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEP_A | 2.26e-68 | 52 | 385 | 35 | 393 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6WQP_A | 1.23e-66 | 38 | 384 | 4 | 354 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 4X0V_A | 2.12e-61 | 51 | 384 | 39 | 393 | Structureof a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_B Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_C Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_D Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_E Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_F Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_G Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_H Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32] |
| 3AYR_A | 4.41e-61 | 48 | 388 | 16 | 362 | GH5endoglucanase EglA from a ruminal fungus [Piromyces rhizinflatus] |
| 5H4R_A | 5.50e-61 | 51 | 384 | 39 | 393 | thecomplex of Glycoside Hydrolase 5 Lichenase from Caldicellulosiruptor sp. F32 E188Q mutant and cellotetraose [Caldicellulosiruptor sp. F32] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 4.66e-65 | 42 | 385 | 30 | 398 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P54937 | 1.46e-58 | 51 | 403 | 38 | 395 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P10477 | 3.12e-57 | 1 | 385 | 1 | 384 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| P23660 | 9.15e-57 | 39 | 371 | 14 | 348 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P20847 | 1.18e-56 | 51 | 384 | 38 | 393 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
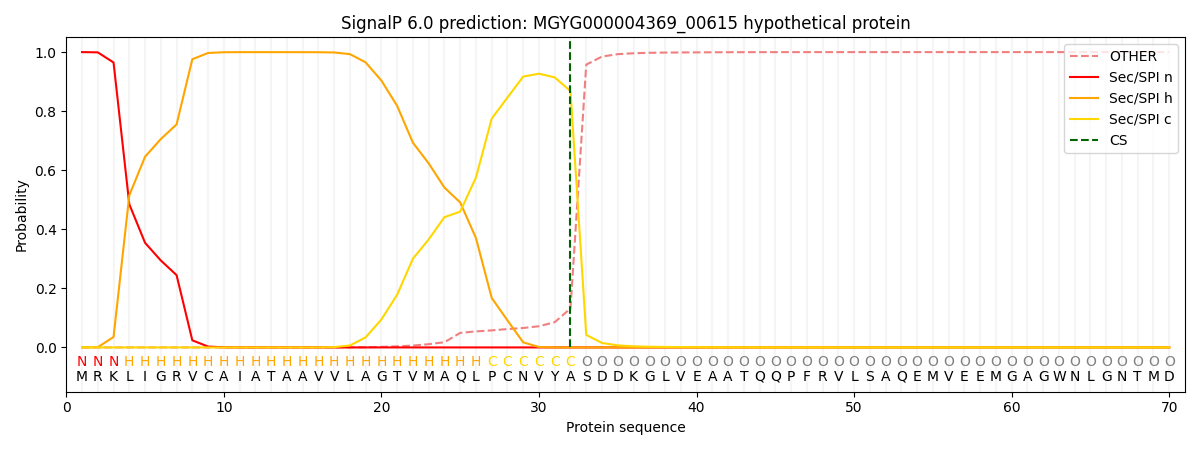
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000436 | 0.998851 | 0.000188 | 0.000191 | 0.000158 | 0.000144 |

