You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004371_01254
You are here: Home > Sequence: MGYG000004371_01254
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002251385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002251385 | |||||||||||
| CAZyme ID | MGYG000004371_01254 | |||||||||||
| CAZy Family | GH143 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5586; End: 7376 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH143 | 19 | 528 | 7e-200 | 0.935251798561151 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13088 | BNR_2 | 2.36e-05 | 264 | 342 | 170 | 252 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| cd15482 | Sialidase_non-viral | 3.28e-04 | 260 | 338 | 95 | 182 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| cd08989 | GH43_XYL-like | 0.009 | 61 | 118 | 52 | 113 | Glycosyl hydrolase family 43, beta-D-xylosidases and arabinofuranosidases. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes that have been annotated as having beta-1,4-xylosidase (beta-D-xylosidase;xylan 1,4-beta-xylosidase; EC 3.2.1.37) activity, including Selenomonas ruminantium beta-D-xylosidase SXA. These are part of an array of hemicellulases that are involved in the final breakdown of plant cell-wall whereby they degrade xylan. They hydrolyze beta-1,4 glycosidic bonds between two xylose units in short xylooligosaccharides. It also includes various GH43 family GH43 arabinofuranosidases (EC 3.2.1.55) including Humicola insolens alpha-L-arabinofuranosidase AXHd3, Bacteroides ovatus alpha-L-arabinofuranosidase (BoGH43, XynB), and the bifunctional Phanerochaete chrysosporium xylosidase/arabinofuranosidase (Xyl;PcXyl). GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYQ36507.1 | 1.43e-253 | 15 | 595 | 18 | 624 |
| AEI49119.1 | 1.59e-252 | 18 | 595 | 21 | 623 |
| QJD77140.1 | 1.48e-250 | 4 | 595 | 5 | 613 |
| QKZ12138.1 | 3.46e-249 | 1 | 595 | 1 | 622 |
| AXE18473.1 | 4.91e-249 | 18 | 595 | 21 | 623 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQS_A | 4.66e-129 | 18 | 594 | 21 | 608 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQR_A | 7.18e-127 | 18 | 594 | 21 | 608 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
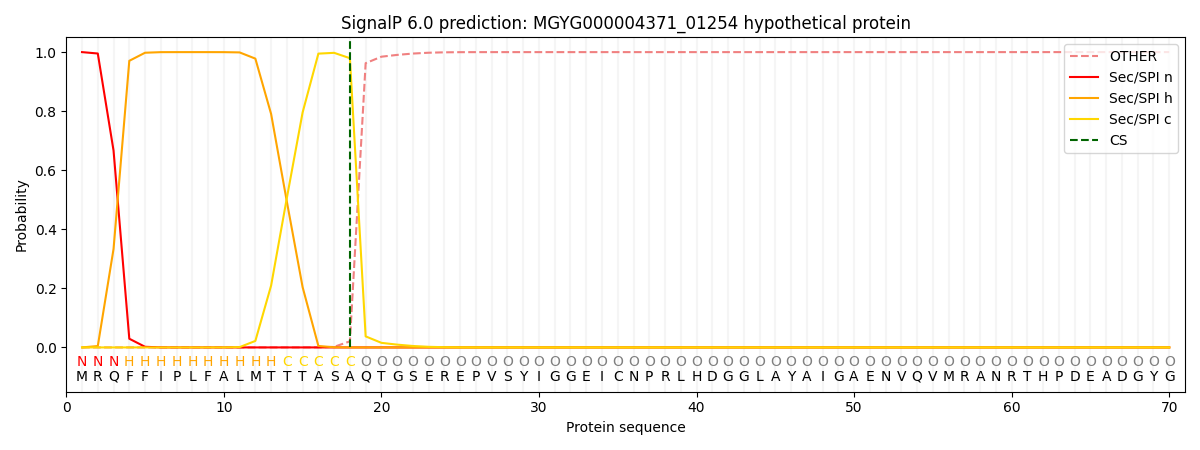
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000236 | 0.999116 | 0.000165 | 0.000154 | 0.000142 | 0.000133 |
