You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004384_00342
You are here: Home > Sequence: MGYG000004384_00342
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; | |||||||||||
| CAZyme ID | MGYG000004384_00342 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17687; End: 20266 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 95 | 276 | 8.7e-53 | 0.9890710382513661 |
| CBM6 | 505 | 631 | 1.6e-28 | 0.9202898550724637 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04084 | CBM6_xylanase-like | 8.39e-37 | 510 | 629 | 2 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| pfam03422 | CBM_6 | 2.88e-27 | 512 | 631 | 1 | 125 | Carbohydrate binding module (family 6). |
| smart00606 | CBD_IV | 1.98e-25 | 504 | 629 | 1 | 129 | Cellulose Binding Domain Type IV. |
| cd14256 | Dockerin_I | 1.15e-14 | 800 | 855 | 1 | 56 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| cd04079 | CBM6_agarase-like | 3.36e-10 | 510 | 631 | 5 | 134 | Carbohydrate Binding Module 6 (CBM6); appended mainly to glycoside hydrolase (GH) family 16 alpha- and beta agarases. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family 16 agarases. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the activity of alpha- and beta-agarase catalytic modules which are involved in the hydrolysis of 1,4-beta-D-galactosidic linkages. These CBM6s bind specifically to the non-reducing end of agarose chains, recognizing only the first repeat of the disaccharide, and directing the appended catalytic modules to areas of the plant cell wall attacked by beta-agarases. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. This family includes three tandem CBM6s from the Saccharophagus degradans agarase Aga86E, and three tandem CBM6s from Vibrio sp. strain PO-303 AgaA; in both these proteins these are appended to a GH16 domain. Vibrio AgaA also contains a Big-2-like protein-protein interaction domain. This family also includes two tandem CBM6s from an endo-type beta-agarase from a deep-sea Microbulbifer-like isolate, which are appended to a GH16 domain, and two of three CBM6s of Alteromonas agarilytica AgaA alpha-agarase, which are appended to a GH96 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU22445.1 | 1.72e-281 | 1 | 796 | 1 | 740 |
| CBL17517.1 | 2.02e-253 | 1 | 855 | 1 | 849 |
| AUO19292.1 | 1.62e-202 | 9 | 804 | 12 | 663 |
| BAV13046.1 | 2.67e-194 | 46 | 502 | 50 | 514 |
| ADL50763.1 | 2.67e-194 | 46 | 502 | 50 | 514 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V4V_A | 9.45e-26 | 506 | 631 | 2 | 129 | CrystalStructure of a Family 6 Carbohydrate-Binding Module from Clostridium cellulolyticum in complex with xylose [Ruminiclostridium cellulolyticum] |
| 1GMM_A | 4.84e-20 | 506 | 634 | 2 | 131 | ChainA, CBM6 [Acetivibrio thermocellus],1UXX_X Chain X, Xylanase U [Acetivibrio thermocellus] |
| 1W9S_A | 1.20e-17 | 508 | 629 | 12 | 138 | ChainA, Bh0236 Protein [Halalkalibacterium halodurans],1W9S_B Chain B, Bh0236 Protein [Halalkalibacterium halodurans],1W9T_A Chain A, Bh0236 Protein [Halalkalibacterium halodurans],1W9T_B Chain B, Bh0236 Protein [Halalkalibacterium halodurans],1W9W_A Chain A, Bh0236 Protein [Halalkalibacterium halodurans] |
| 1O8P_A | 5.23e-17 | 506 | 631 | 24 | 148 | Unboundstructure of CsCBM6-3 from Clostridium stercorarium [Thermoclostridium stercorarium] |
| 1NAE_A | 8.03e-17 | 506 | 631 | 41 | 165 | Structureof CsCBM6-3 from Clostridium stercorarium in complex with xylotriose [Thermoclostridium stercorarium],1O8S_A Structure of CsCBM6-3 from Clostridium stercorarium in complex with cellobiose [Thermoclostridium stercorarium],1OD3_A Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose [Thermoclostridium stercorarium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 6.91e-44 | 46 | 489 | 22 | 418 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 8.08e-43 | 46 | 489 | 22 | 418 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| A1DPF0 | 3.25e-41 | 46 | 489 | 23 | 419 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| Q4WL88 | 4.41e-41 | 46 | 489 | 23 | 419 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| B0XMA2 | 4.41e-41 | 46 | 489 | 23 | 419 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
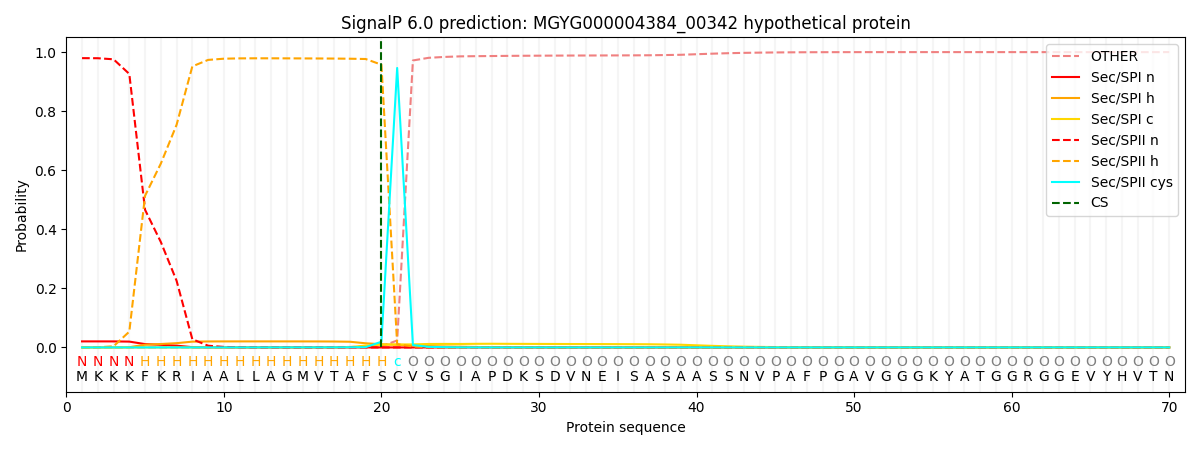
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000080 | 0.019654 | 0.980211 | 0.000018 | 0.000024 | 0.000016 |
