You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004392_01541
You are here: Home > Sequence: MGYG000004392_01541
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Eubacterium_R sp900539325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Eubacterium_R; Eubacterium_R sp900539325 | |||||||||||
| CAZyme ID | MGYG000004392_01541 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57355; End: 59994 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 51 | 865 | 5.3e-117 | 0.9847645429362881 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd20789 | Cas13d | 0.008 | 195 | 339 | 3 | 193 | Class 2 type VI-D CRISPR-associated RNA-guided ribonuclease Cas13d. CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) adaptive immune systems defend microbes against foreign nucleic acids via RNA-guided endonucleases. These systems are divided into two classes: class 1 systems utilize multiple Cas proteins and CRISPR RNA (crRNA) to form an effector complex while class 2 systems employ a large, single effector with crRNA to mediate interference. Class 2 type VI CRISPR-Cas13 systems use a single enzyme to target RNA using a programmable crRNA guide and are divided into four subtypes based on the identity of the Cas13 protein (Cas13a-d). The Cas13 proteins are capable of both, pre-crRNA processing and target RNA cleavage, which protect the host from phage attacks. Once bound to a target RNA, their non-specific RNase activity is activated. Cas13d enzymes are 20-30% smaller than other Cas13 subtypes, which enable flexible packaging into size-constrained therapeutic viral vectors such as adeno-associated virus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19599.1 | 1.15e-242 | 44 | 879 | 7 | 830 |
| QYR23103.1 | 4.34e-231 | 41 | 874 | 13 | 865 |
| AIQ51360.1 | 2.64e-227 | 41 | 850 | 13 | 852 |
| AIQ45759.1 | 1.01e-225 | 41 | 850 | 13 | 852 |
| AFH60346.1 | 1.60e-223 | 41 | 861 | 13 | 861 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UFC_A | 6.68e-20 | 346 | 858 | 281 | 741 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 2RDY_A | 1.84e-13 | 346 | 858 | 281 | 752 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 7.88e-13 | 324 | 837 | 315 | 792 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 1.54e-11 | 153 | 838 | 137 | 757 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
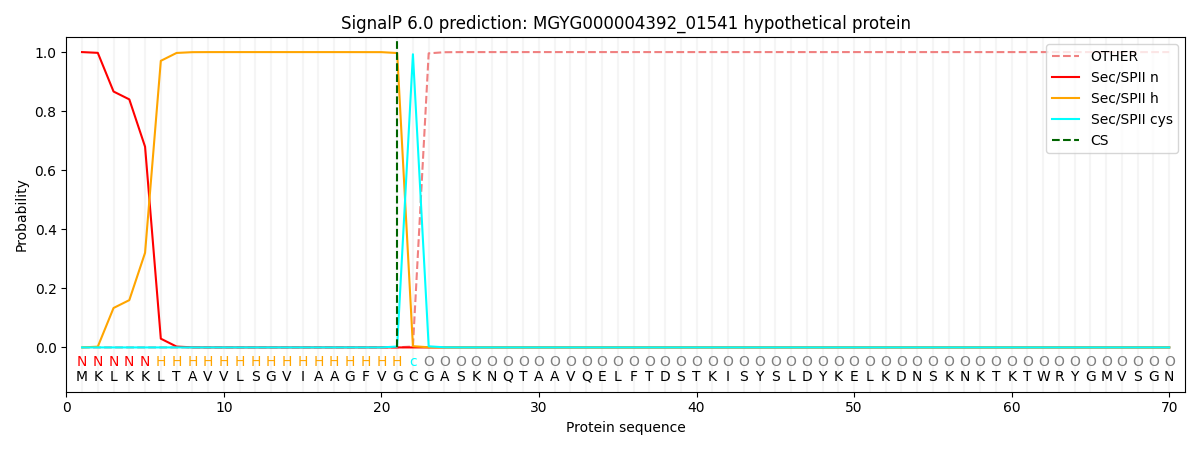
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000068 | 0.000000 | 0.000000 | 0.000000 |
