You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004397_00040
You are here: Home > Sequence: MGYG000004397_00040
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
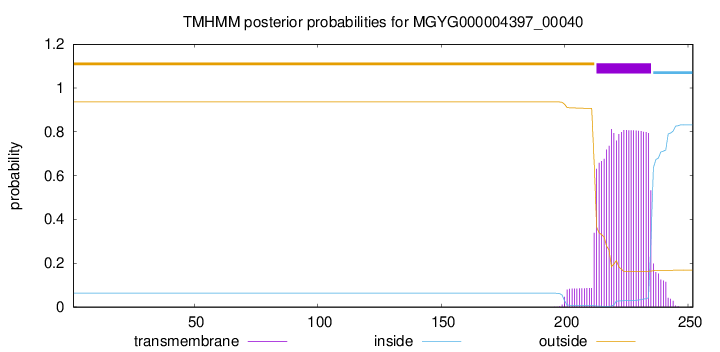
TMHMM annotations
Basic Information help
| Species | Caecibacter sp900549385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Megasphaeraceae; Caecibacter; Caecibacter sp900549385 | |||||||||||
| CAZyme ID | MGYG000004397_00040 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42191; End: 42949 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02511 | Beta4Glucosyltransferase | 6.92e-75 | 4 | 229 | 2 | 229 | UDP-glucose LOS-beta-1,4 glucosyltransferase is required for biosynthesis of lipooligosaccharide. UDP-glucose: lipooligosaccharide (LOS) beta-1-4-glucosyltransferase catalyzes the addition of the first residue, glucose, of the lacto-N-neotetrase structure to HepI of the LOS inner core. LOS is the major constituent of the outer leaflet of the outer membrane of gram-positive bacteria. It consists of a short oligosaccharide chain of variable composition (alpha chain) attached to a branched inner core which is lined in turn to lipid A. Beta 1,4 glucosyltransferase is required to attach the alpha chain to the inner core. |
| COG0463 | WcaA | 5.42e-19 | 1 | 252 | 2 | 263 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| cd00761 | Glyco_tranf_GTA_type | 3.20e-15 | 6 | 117 | 1 | 119 | Glycosyltransferase family A (GT-A) includes diverse families of glycosyl transferases with a common GT-A type structural fold. Glycosyltransferases (GTs) are enzymes that synthesize oligosaccharides, polysaccharides, and glycoconjugates by transferring the sugar moiety from an activated nucleotide-sugar donor to an acceptor molecule, which may be a growing oligosaccharide, a lipid, or a protein. Based on the stereochemistry of the donor and acceptor molecules, GTs are classified as either retaining or inverting enzymes. To date, all GT structures adopt one of two possible folds, termed GT-A fold and GT-B fold. This hierarchy includes diverse families of glycosyl transferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. The majority of the proteins in this superfamily are Glycosyltransferase family 2 (GT-2) proteins. But it also includes families GT-43, GT-6, GT-8, GT13 and GT-7; which are evolutionarily related to GT-2 and share structure similarities. |
| pfam00535 | Glycos_transf_2 | 3.98e-15 | 6 | 92 | 2 | 96 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
| cd02522 | GT_2_like_a | 1.95e-12 | 4 | 95 | 1 | 93 | GT_2_like_a represents a glycosyltransferase family-2 subfamily with unknown function. Glycosyltransferase family 2 (GT-2) subfamily of unknown function. GT-2 includes diverse families of glycosyltransferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. These are enzymes that catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. Glycosyltransferases have been classified into more than 90 distinct sequence based families. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVO28104.1 | 1.14e-143 | 1 | 252 | 1 | 252 |
| ALG42578.1 | 1.33e-142 | 1 | 252 | 1 | 252 |
| CCC73969.1 | 1.74e-140 | 1 | 251 | 1 | 251 |
| AVO75335.1 | 1.74e-140 | 1 | 251 | 1 | 251 |
| AXB82128.1 | 2.71e-110 | 1 | 249 | 1 | 249 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MSK_A | 2.01e-09 | 3 | 150 | 67 | 211 | ChainA, Glyco_trans_2-like domain-containing protein [Bacillus thuringiensis serovar andalousiensis BGSC 4AW1],7MSK_B Chain B, Glyco_trans_2-like domain-containing protein [Bacillus thuringiensis serovar andalousiensis BGSC 4AW1] |
| 7MSP_A | 7.32e-09 | 8 | 150 | 60 | 200 | ChainA, SPbeta prophage-derived glycosyltransferase SunS [Bacillus subtilis subsp. subtilis str. 168],7MSP_B Chain B, SPbeta prophage-derived glycosyltransferase SunS [Bacillus subtilis subsp. subtilis str. 168] |
| 7MSN_A | 8.72e-09 | 8 | 150 | 60 | 200 | ChainA, SPbeta prophage-derived glycosyltransferase SunS [Bacillus subtilis subsp. subtilis str. 168],7MSN_B Chain B, SPbeta prophage-derived glycosyltransferase SunS [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P44029 | 2.71e-40 | 1 | 250 | 1 | 253 | Uncharacterized glycosyltransferase HI_0653 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_0653 PE=3 SV=1 |
| Q9XC90 | 4.24e-40 | 2 | 246 | 3 | 248 | Lipopolysaccharide core biosynthesis glycosyltransferase WaaE OS=Klebsiella pneumoniae OX=573 GN=waaE PE=3 SV=1 |
| Q54435 | 1.61e-31 | 3 | 246 | 6 | 250 | Lipopolysaccharide core biosynthesis glycosyltransferase KdtX OS=Serratia marcescens OX=615 GN=kdtX PE=3 SV=1 |
| Q68XF1 | 8.69e-21 | 1 | 249 | 1 | 260 | Uncharacterized glycosyltransferase RT0209 OS=Rickettsia typhi (strain ATCC VR-144 / Wilmington) OX=257363 GN=RT0209 PE=3 SV=1 |
| O05944 | 6.16e-20 | 1 | 251 | 1 | 262 | Uncharacterized glycosyltransferase RP128 OS=Rickettsia prowazekii (strain Madrid E) OX=272947 GN=RP218 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
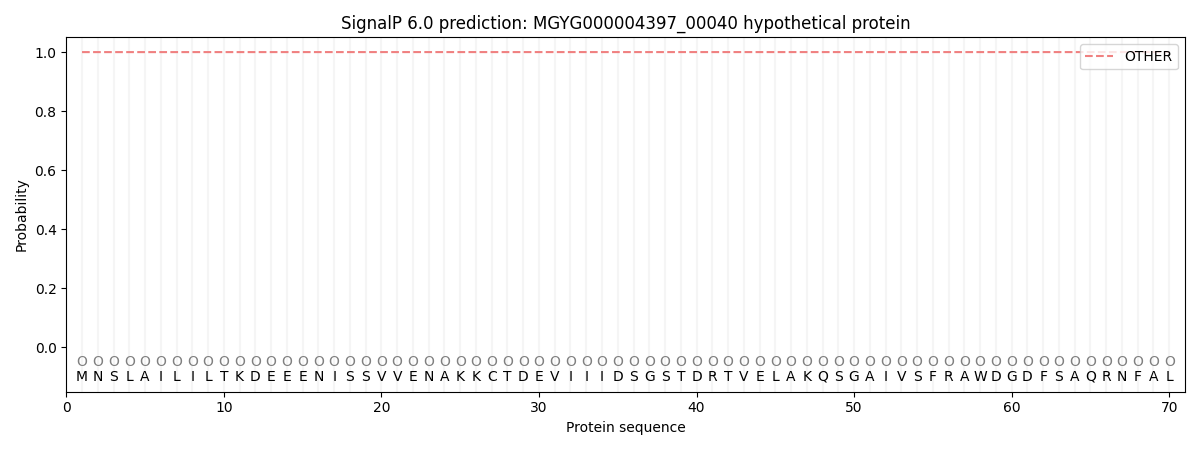
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000046 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |