You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004402_01357
You are here: Home > Sequence: MGYG000004402_01357
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000432515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000432515 | |||||||||||
| CAZyme ID | MGYG000004402_01357 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47925; End: 53981 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 761 | 1072 | 9.6e-116 | 0.990228013029316 |
| GH20 | 1636 | 1995 | 6.8e-108 | 0.9584569732937686 |
| GH78 | 278 | 715 | 3.3e-84 | 0.8888888888888888 |
| CBM67 | 26 | 189 | 3.3e-32 | 0.8920454545454546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06563 | GH20_chitobiase-like | 2.88e-165 | 1637 | 2008 | 2 | 357 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam01301 | Glyco_hydro_35 | 7.60e-149 | 760 | 1073 | 1 | 316 | Glycosyl hydrolases family 35. |
| pfam00728 | Glyco_hydro_20 | 1.05e-118 | 1637 | 1995 | 2 | 345 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| COG3525 | Chb | 9.44e-100 | 1579 | 2000 | 201 | 620 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
| cd06570 | GH20_chitobiase-like_1 | 1.60e-80 | 1638 | 2000 | 3 | 303 | A functionally uncharacterized subgroup of the Glycosyl hydrolase family 20 (GH20) catalytic domain found in proteins similar to the chitobiase of Serratia marcescens, a beta-N-1,4-acetylhexosaminidase that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This subgroup lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANU56223.1 | 0.0 | 753 | 1500 | 28 | 778 |
| QQR18938.1 | 0.0 | 753 | 1500 | 28 | 778 |
| QRM98976.1 | 0.0 | 753 | 1500 | 28 | 778 |
| ALJ48378.1 | 0.0 | 753 | 1500 | 28 | 778 |
| QRQ55220.1 | 0.0 | 753 | 1500 | 28 | 778 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EON_A | 0.0 | 751 | 1498 | 25 | 777 | GalactanaseBT0290 [Bacteroides thetaiotaomicron VPI-5482] |
| 3D3A_A | 6.83e-274 | 751 | 1348 | 5 | 602 | Crystalstructure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4MAD_A | 6.31e-138 | 761 | 1335 | 24 | 593 | ChainA, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
| 4E8C_A | 2.27e-112 | 760 | 1343 | 9 | 595 | Crystalstructure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8C_B Crystal structure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8D_A Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4],4E8D_B Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4] |
| 3WF3_A | 9.35e-108 | 740 | 1319 | 23 | 622 | Crystalstructure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_B Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_C Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_D Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF4_A Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_B Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_C Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_D Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48982 | 6.94e-116 | 752 | 1302 | 28 | 573 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
| P16278 | 1.25e-106 | 747 | 1319 | 29 | 621 | Beta-galactosidase OS=Homo sapiens OX=9606 GN=GLB1 PE=1 SV=2 |
| Q60HF6 | 6.59e-106 | 753 | 1319 | 33 | 621 | Beta-galactosidase OS=Macaca fascicularis OX=9541 GN=GLB1 PE=2 SV=1 |
| Q95LV1 | 6.58e-105 | 761 | 1336 | 39 | 625 | Beta-galactosidase-1-like protein OS=Macaca fascicularis OX=9541 GN=GLB1L PE=2 SV=1 |
| Q5R7P4 | 6.65e-105 | 747 | 1319 | 29 | 621 | Beta-galactosidase OS=Pongo abelii OX=9601 GN=GLB1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
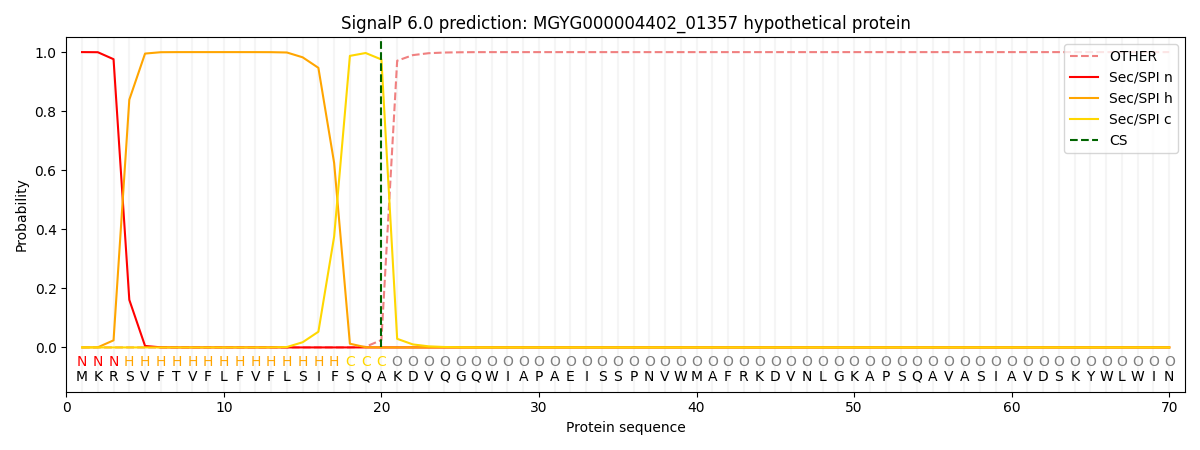
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000270 | 0.999033 | 0.000182 | 0.000182 | 0.000162 | 0.000145 |
