You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004414_01731
You are here: Home > Sequence: MGYG000004414_01731
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
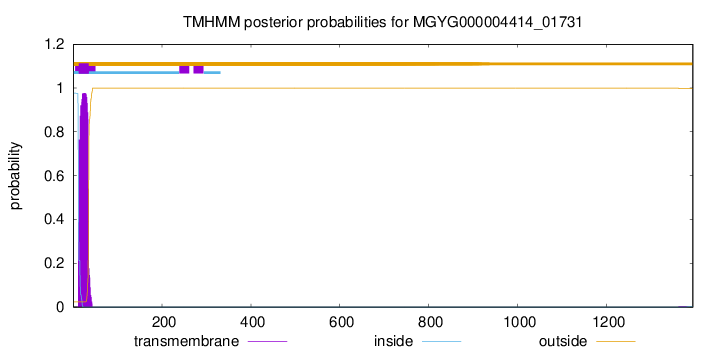
TMHMM annotations
Basic Information help
| Species | Agathobacter sp900549895 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter sp900549895 | |||||||||||
| CAZyme ID | MGYG000004414_01731 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15225; End: 19409 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 420 | 755 | 7.3e-60 | 0.9273927392739274 |
| CBM23 | 924 | 1079 | 2e-33 | 0.8827160493827161 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 5.62e-38 | 420 | 753 | 1 | 288 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 3.97e-16 | 556 | 749 | 155 | 340 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 6.88e-04 | 920 | 1048 | 4 | 128 | Carbohydrate binding domain (family 11). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL07467.1 | 2.26e-233 | 82 | 1114 | 78 | 1065 |
| VCV20359.1 | 1.47e-232 | 60 | 1114 | 57 | 1065 |
| EEV00397.1 | 2.05e-232 | 60 | 1114 | 69 | 1077 |
| CBL14297.1 | 5.72e-232 | 60 | 1114 | 57 | 1065 |
| CBK83362.1 | 2.41e-231 | 77 | 1110 | 434 | 1430 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 1.09e-52 | 418 | 793 | 50 | 426 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 1J9Y_A | 1.01e-49 | 417 | 749 | 9 | 324 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1R7O_A | 1.29e-49 | 417 | 749 | 19 | 334 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
| 2WHM_A | 1.86e-49 | 417 | 749 | 9 | 324 | Cellvibriojaponicus Man26A E121A and E320G double mutant in complex with mannobiose [Cellvibrio japonicus] |
| 1GW1_A | 5.02e-49 | 417 | 749 | 5 | 320 | Substratedistortion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 1.39e-48 | 417 | 749 | 47 | 362 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 5.58e-42 | 417 | 1099 | 40 | 697 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P49425 | 5.34e-19 | 373 | 669 | 103 | 383 | Mannan endo-1,4-beta-mannosidase OS=Rhodothermus marinus (strain ATCC 43812 / DSM 4252 / R-10) OX=518766 GN=manA PE=1 SV=3 |
| P55278 | 5.05e-14 | 420 | 667 | 30 | 272 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| O05512 | 6.88e-14 | 414 | 681 | 26 | 284 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
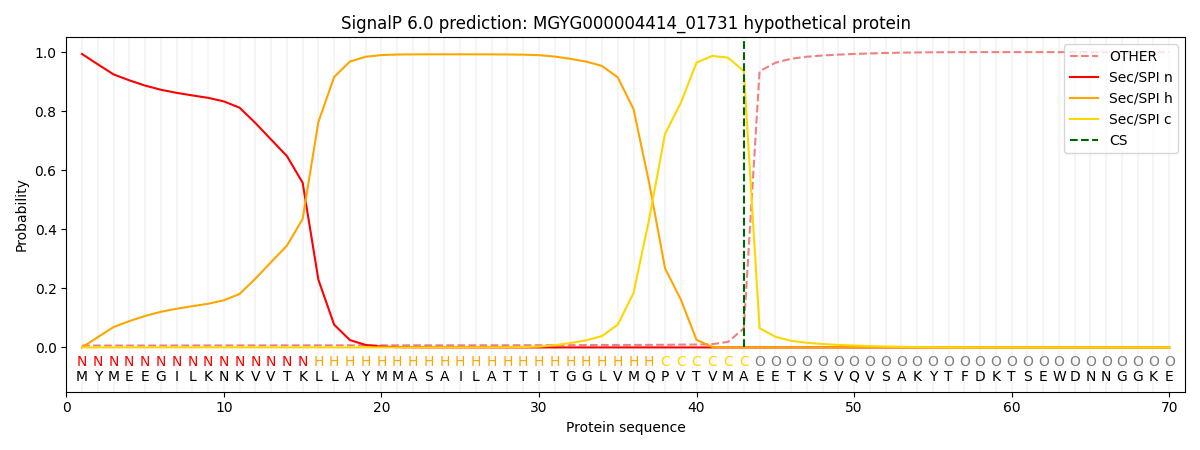
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007737 | 0.990926 | 0.000416 | 0.000435 | 0.000241 | 0.000222 |