You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004428_00935
You are here: Home > Sequence: MGYG000004428_00935
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; | |||||||||||
| CAZyme ID | MGYG000004428_00935 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25298; End: 26737 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 44 | 245 | 4.5e-54 | 0.8482142857142857 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11790 | Glyco_hydro_cc | 6.77e-58 | 45 | 281 | 17 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU93826.1 | 6.27e-61 | 22 | 412 | 51 | 427 |
| ARU26557.1 | 1.12e-47 | 23 | 412 | 115 | 483 |
| QKX17146.1 | 1.38e-45 | 20 | 413 | 738 | 1104 |
| AYN96242.1 | 3.00e-45 | 20 | 251 | 29 | 261 |
| ABD82184.1 | 3.45e-45 | 22 | 415 | 735 | 1105 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UAV_A | 3.85e-41 | 22 | 243 | 2 | 226 | Crystalstructure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Pseudomonas viridiflava (PvGH128_II) [Pseudomonas viridiflava] |
| 6UAW_A | 7.03e-41 | 22 | 243 | 24 | 248 | Crystalstructure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Pseudomonas viridiflava (PvGH128_II) in complex with laminaritriose [Pseudomonas viridiflava] |
| 6UAX_A | 4.60e-34 | 25 | 251 | 95 | 322 | Crystalstructure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Sorangium cellulosum (ScGH128_II) [Sorangium cellulosum So ce56],6UAX_B Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Sorangium cellulosum (ScGH128_II) [Sorangium cellulosum So ce56] |
| 6UAQ_A | 3.00e-21 | 25 | 257 | 25 | 255 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
| 6UFL_A | 7.57e-21 | 25 | 257 | 25 | 255 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
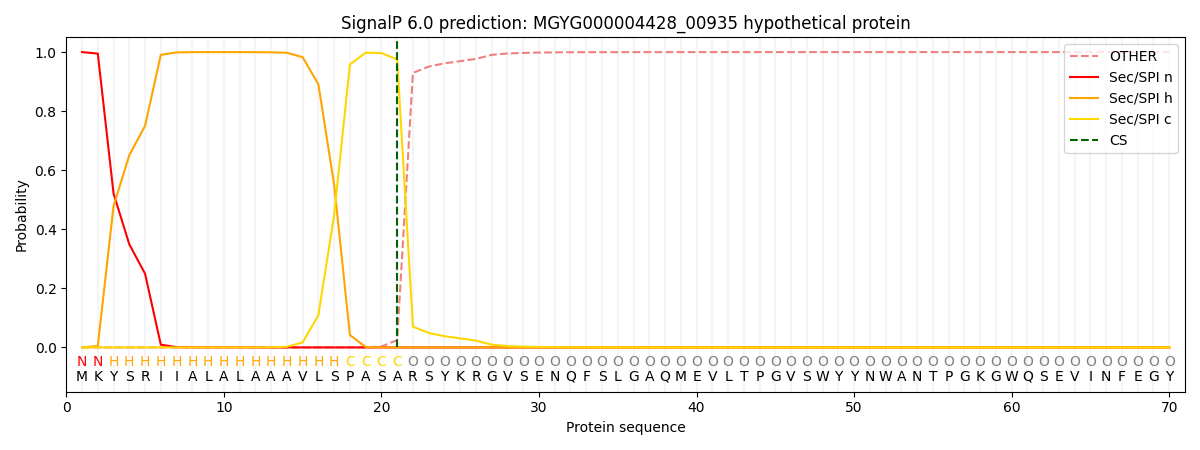
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000468 | 0.998475 | 0.000301 | 0.000276 | 0.000232 | 0.000208 |
