You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004429_01746
You are here: Home > Sequence: MGYG000004429_01746
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
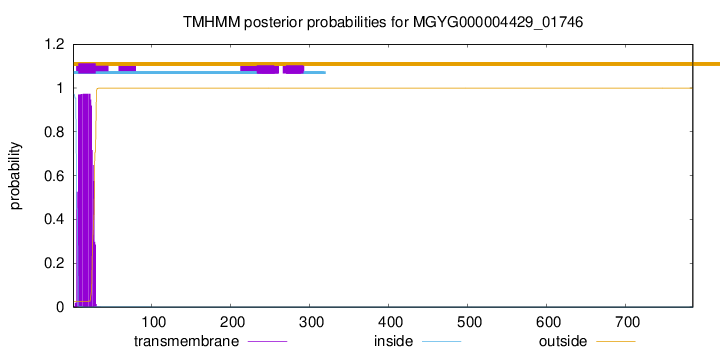
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; ; | |||||||||||
| CAZyme ID | MGYG000004429_01746 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 502; End: 2859 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 84 | 275 | 7.9e-73 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17957 | Big_7 | 3.35e-14 | 468 | 532 | 1 | 67 | Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
| COG3866 | PelB | 2.80e-09 | 41 | 282 | 49 | 279 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 2.44e-07 | 82 | 271 | 1 | 179 | Amb_all domain. |
| pfam07081 | DUF1349 | 1.59e-06 | 599 | 762 | 31 | 149 | Protein of unknown function (DUF1349). This family consists of several hypothetical bacterial proteins but contains one sequence from Saccharomyces cerevisiae. Members of this family are typically around 200 residues in length. The function of this family is unknown. |
| pfam00544 | Pec_lyase_C | 1.48e-04 | 122 | 259 | 57 | 197 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYR21100.1 | 5.32e-223 | 26 | 784 | 45 | 832 |
| AWV33235.1 | 9.14e-222 | 22 | 784 | 48 | 839 |
| AFH63199.1 | 5.81e-221 | 23 | 785 | 37 | 761 |
| AFC30874.1 | 5.81e-221 | 23 | 785 | 37 | 761 |
| AIQ73887.1 | 1.71e-220 | 22 | 785 | 42 | 834 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PDG_A | 1.08e-09 | 468 | 551 | 2 | 86 | ChainA, Fibronectin(III)-like module [Acetivibrio thermocellus ATCC 27405] |
| 3PDD_A | 8.03e-09 | 468 | 551 | 2 | 86 | ChainA, Glycoside hydrolase, family 9 [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5B297 | 8.58e-60 | 25 | 457 | 18 | 411 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 1.44e-57 | 11 | 457 | 4 | 414 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| B0XMA2 | 1.85e-55 | 23 | 457 | 17 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| B8NQQ7 | 2.49e-55 | 29 | 457 | 22 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q4WL88 | 2.55e-55 | 23 | 457 | 17 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000270 | 0.999080 | 0.000178 | 0.000168 | 0.000153 | 0.000135 |