You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004430_01151
You are here: Home > Sequence: MGYG000004430_01151
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000004430_01151 | |||||||||||
| CAZy Family | GH53 | |||||||||||
| CAZyme Description | Arabinogalactan endo-beta-1,4-galactanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 23979; End: 25025 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 34 | 320 | 3e-90 | 0.8947368421052632 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07745 | Glyco_hydro_53 | 1.99e-91 | 34 | 319 | 3 | 301 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| COG3867 | GanB | 3.30e-84 | 28 | 318 | 36 | 345 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| cd11330 | AmyAc_OligoGlu | 0.005 | 64 | 127 | 55 | 139 | Alpha amylase catalytic domain found in oligo-1,6-glucosidase (also called isomaltase; sucrase-isomaltase; alpha-limit dextrinase) and related proteins. Oligo-1,6-glucosidase (EC 3.2.1.10) hydrolyzes the alpha-1,6-glucosidic linkage of isomalto-oligosaccharides, pannose, and dextran. Unlike alpha-1,4-glucosidases (EC 3.2.1.20), it fails to hydrolyze the alpha-1,4-glucosidic bonds of maltosaccharides. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIF26228.1 | 6.87e-133 | 1 | 341 | 1 | 333 |
| BCS85804.1 | 1.75e-123 | 15 | 340 | 9 | 333 |
| AGB28883.1 | 5.70e-119 | 16 | 345 | 10 | 337 |
| SCM59778.1 | 2.51e-106 | 14 | 341 | 13 | 359 |
| QJR97948.1 | 1.19e-97 | 13 | 344 | 1 | 354 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GPA_A | 5.08e-91 | 29 | 341 | 5 | 314 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482],6GPA_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482] |
| 6GP5_A | 1.69e-90 | 29 | 341 | 41 | 350 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],6GP5_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4BF7_A | 4.55e-41 | 34 | 317 | 23 | 317 | Emericillanidulans endo-beta-1,4-galactanase [Aspergillus nidulans] |
| 7OSK_A | 2.72e-40 | 37 | 317 | 51 | 356 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 1R8L_A | 2.38e-39 | 29 | 317 | 22 | 324 | Thestructure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1R8L_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1UR0_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR0_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],2CCR_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2CCR_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48843 | 1.78e-51 | 29 | 317 | 4 | 312 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| A1D3T4 | 4.38e-44 | 34 | 317 | 27 | 323 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=galA PE=3 SV=1 |
| B0XPR3 | 3.30e-43 | 13 | 317 | 8 | 323 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=galA PE=3 SV=2 |
| Q9Y7F8 | 1.11e-42 | 34 | 317 | 22 | 317 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus tubingensis OX=5068 GN=galA PE=2 SV=1 |
| Q4WJ80 | 1.27e-42 | 13 | 317 | 8 | 323 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=galA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
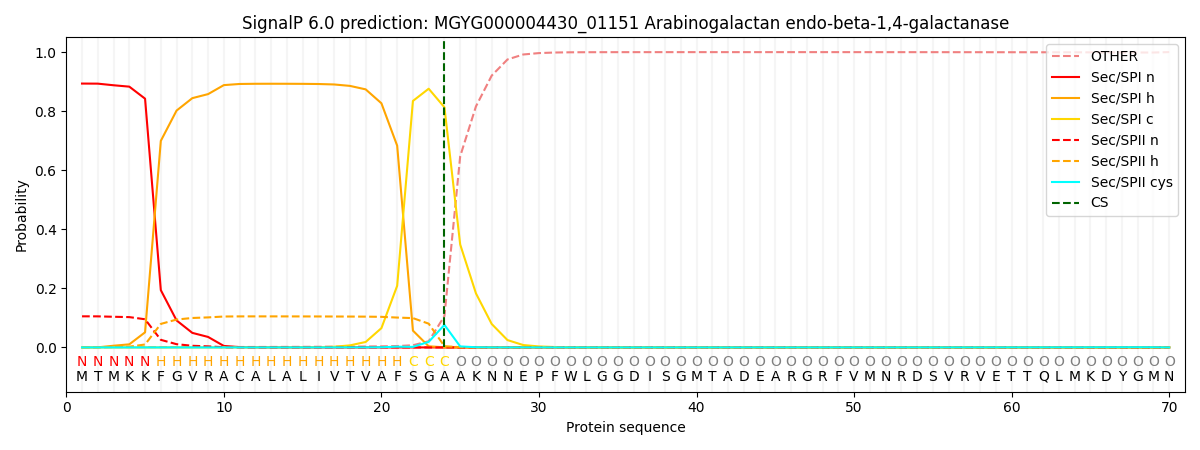
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004112 | 0.884855 | 0.109925 | 0.000350 | 0.000354 | 0.000368 |
