You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004430_01448
You are here: Home > Sequence: MGYG000004430_01448
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000004430_01448 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 101763; End: 105278 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 241 | 852 | 2.6e-91 | 0.819047619047619 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 1.70e-37 | 244 | 854 | 33 | 501 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 3.82e-17 | 486 | 576 | 1 | 95 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 5.49e-17 | 486 | 563 | 1 | 79 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam02018 | CBM_4_9 | 5.00e-05 | 105 | 241 | 8 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ60076.1 | 1.86e-227 | 23 | 852 | 25 | 837 |
| QUT88785.1 | 3.38e-224 | 9 | 853 | 10 | 836 |
| ALJ60214.1 | 7.48e-223 | 9 | 853 | 10 | 836 |
| QJR66308.1 | 3.31e-197 | 1 | 855 | 1 | 826 |
| QJR70571.1 | 3.31e-197 | 1 | 855 | 1 | 826 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 8.20e-83 | 32 | 563 | 8 | 518 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 2VRQ_A | 2.78e-09 | 260 | 459 | 50 | 222 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 6ZT6_A | 4.84e-09 | 260 | 459 | 50 | 222 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZTA_A | 4.84e-09 | 260 | 459 | 50 | 222 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZT8_A | 1.46e-08 | 260 | 410 | 50 | 178 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 3.25e-131 | 10 | 852 | 14 | 823 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 2.82e-78 | 20 | 580 | 31 | 559 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 8.15e-77 | 20 | 580 | 32 | 558 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q8NK90 | 8.26e-70 | 34 | 563 | 35 | 526 | Alpha-L-arabinofuranosidase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=abfA PE=1 SV=2 |
| Q96X54 | 1.53e-69 | 34 | 563 | 35 | 526 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus awamori OX=105351 GN=abfA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
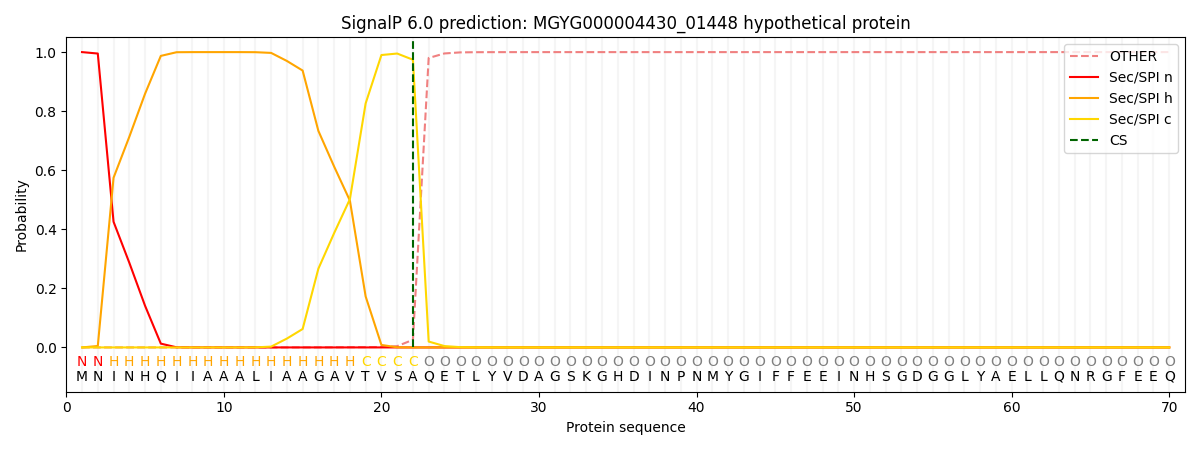
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000228 | 0.999147 | 0.000154 | 0.000161 | 0.000153 | 0.000138 |
