You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004430_01631
You are here: Home > Sequence: MGYG000004430_01631
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000004430_01631 | |||||||||||
| CAZy Family | GH89 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26595; End: 29957 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH89 | 463 | 1107 | 2.6e-236 | 0.9773755656108597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05089 | NAGLU | 5.44e-179 | 521 | 850 | 1 | 333 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| pfam12972 | NAGLU_C | 1.64e-84 | 858 | 1113 | 1 | 257 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| pfam12971 | NAGLU_N | 9.21e-28 | 428 | 506 | 1 | 81 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This N-terminal domain has an alpha-beta fold. |
| pfam00328 | His_Phos_2 | 1.28e-11 | 73 | 396 | 10 | 353 | Histidine phosphatase superfamily (branch 2). The histidine phosphatase superfamily is so named because catalysis centers on a conserved His residue that is transiently phosphorylated during the catalytic cycle. Other conserved residues contribute to a 'phosphate pocket' and interact with the phospho group of substrate before, during and after its transfer to the His residue. Structure and sequence analyses show that different families contribute different additional residues to the 'phosphate pocket' and, more surprisingly, differ in the position, in sequence and in three dimensions, of a catalytically essential acidic residue. The superfamily may be divided into two main branches.The smaller branch 2 contains predominantly eukaryotic proteins. The catalytic functions in members include phytase, glucose-1-phosphatase and multiple inositol polyphosphate phosphatase. The in vivo roles of the mammalian acid phosphatases in branch 2 are not fully understood, although activity against lysophosphatidic acid and tyrosine-phosphorylated proteins has been demonstrated. |
| cd07061 | HP_HAP_like | 1.16e-08 | 124 | 378 | 17 | 222 | Histidine phosphatase domain found in histidine acid phosphatases and phytases; contains a His residue which is phosphorylated during the reaction. Catalytic domain of HAP (histidine acid phosphatases) and phytases (myo-inositol hexakisphosphate phosphohydrolases). The conserved catalytic core of this domain contains a His residue which is phosphorylated in the reaction. Functions in this subgroup include roles in metabolism, signaling, or regulation, for example Escherichia coli glucose-1-phosphatase functions to scavenge glucose from glucose-1-phosphate and the signaling molecules inositol 1,3,4,5,6-pentakisphosphate (InsP5) and inositol hexakisphosphate (InsP6) are in vivo substrates for eukaryotic multiple inositol polyphosphate phosphatase 1 (Minpp1). Phytases scavenge phosphate from extracellular sources and are added to animal feed while prostatic acid phosphatase (PAP) has been used for many years as a serum marker for prostate cancer. Recently PAP has been shown in mouse models to suppress pain by functioning as an ecto-5prime-nucleotidase. In vivo it dephosphorylates extracellular adenosine monophosphate (AMP) generating adenosine,and leading to the activation of A1-adenosine receptors in dorsal spinal cord. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50280.1 | 1.07e-301 | 426 | 1102 | 18 | 696 |
| QRO24616.1 | 4.29e-301 | 428 | 1120 | 20 | 715 |
| QTO27209.1 | 8.30e-288 | 428 | 1102 | 21 | 698 |
| QCQ31494.1 | 1.66e-287 | 428 | 1102 | 21 | 698 |
| QCQ35767.1 | 1.66e-287 | 428 | 1102 | 21 | 698 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VC9_A | 1.74e-152 | 428 | 1099 | 172 | 850 | Family89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 2.16e-152 | 428 | 1099 | 180 | 858 | ChainA, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
| 4A4A_A | 2.44e-151 | 428 | 1099 | 195 | 873 | CpGH89(E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
| 4XWH_A | 2.89e-114 | 468 | 1115 | 54 | 706 | Crystalstructure of the human N-acetyl-alpha-glucosaminidase [Homo sapiens] |
| 7R5Y_A | 1.16e-49 | 32 | 430 | 7 | 406 | ChainA, Histidine acid phosphatase [Prevotella sp. CAG:617],7R5Y_B Chain B, Histidine acid phosphatase [Prevotella sp. CAG:617],7R5Y_C Chain C, Histidine acid phosphatase [Prevotella sp. CAG:617],7R5Y_D Chain D, Histidine acid phosphatase [Prevotella sp. CAG:617],7R5Y_E Chain E, Histidine acid phosphatase [Prevotella sp. CAG:617],7R5Y_F Chain F, Histidine acid phosphatase [Prevotella sp. CAG:617] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FNA3 | 8.73e-124 | 433 | 1113 | 69 | 797 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
| P54802 | 2.89e-113 | 468 | 1115 | 77 | 729 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
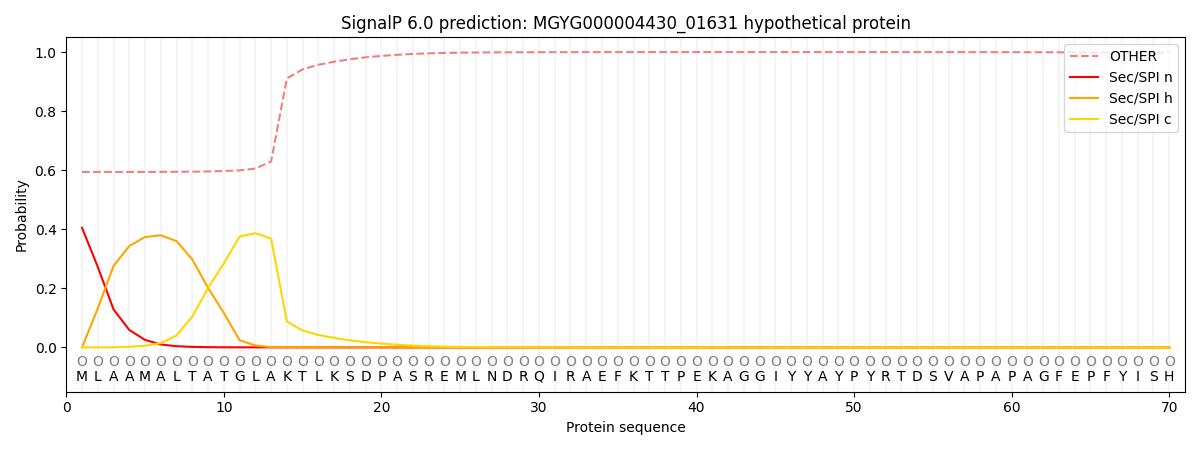
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.608274 | 0.389194 | 0.001086 | 0.000814 | 0.000343 | 0.000294 |
