You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004445_01023
You are here: Home > Sequence: MGYG000004445_01023
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
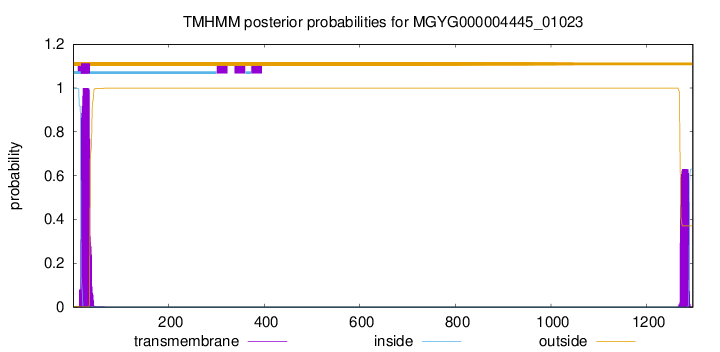
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; ; | |||||||||||
| CAZyme ID | MGYG000004445_01023 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30006; End: 33899 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 536 | 786 | 1.3e-21 | 0.5907692307692308 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 4.52e-21 | 478 | 784 | 76 | 369 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam09479 | Flg_new | 3.00e-10 | 1175 | 1240 | 1 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| pfam00295 | Glyco_hydro_28 | 1.01e-04 | 592 | 788 | 48 | 214 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| pfam12708 | Pectate_lyase_3 | 0.005 | 484 | 521 | 2 | 37 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23464.1 | 6.38e-169 | 221 | 925 | 71 | 752 |
| QCD41820.1 | 2.04e-108 | 215 | 924 | 196 | 884 |
| QGQ94767.1 | 7.11e-41 | 481 | 981 | 34 | 533 |
| ADB48560.1 | 2.77e-38 | 481 | 953 | 25 | 486 |
| ADB48561.1 | 1.93e-37 | 481 | 936 | 25 | 460 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MXN_A | 1.31e-12 | 491 | 691 | 27 | 210 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
| 3JUR_A | 5.47e-07 | 490 | 715 | 32 | 267 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
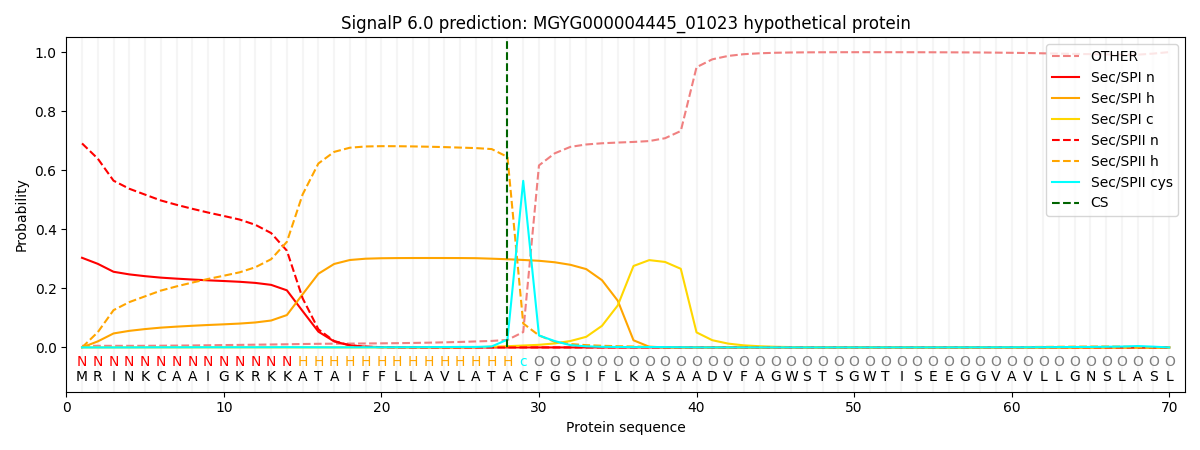
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007076 | 0.292615 | 0.698942 | 0.000638 | 0.000364 | 0.000349 |