You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004461_02557
You are here: Home > Sequence: MGYG000004461_02557
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; ; | |||||||||||
| CAZyme ID | MGYG000004461_02557 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25413; End: 30002 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 99 | 487 | 1.4e-51 | 0.4441489361702128 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.93e-26 | 90 | 469 | 51 | 430 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.54e-15 | 104 | 468 | 67 | 445 | beta-D-glucuronidase; Provisional |
| PRK09525 | lacZ | 2.69e-13 | 106 | 483 | 124 | 500 | beta-galactosidase. |
| pfam00703 | Glyco_hydro_2 | 1.04e-12 | 208 | 317 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 1.89e-12 | 106 | 473 | 113 | 477 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHF94339.1 | 5.97e-253 | 37 | 1528 | 54 | 1609 |
| QUH31336.1 | 4.91e-44 | 108 | 1031 | 65 | 1022 |
| AIQ71089.1 | 1.12e-36 | 114 | 1115 | 89 | 1169 |
| AIQ48916.1 | 4.57e-36 | 36 | 910 | 8 | 973 |
| AIQ54360.1 | 2.42e-35 | 280 | 910 | 374 | 1029 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DYM_A | 6.60e-18 | 106 | 483 | 123 | 499 | ChainA, Beta-galactosidase [Escherichia coli K-12],3DYM_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYM_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYM_D Chain D, Beta-galactosidase [Escherichia coli K-12],3E1F_1 Chain 1, Beta-galactosidase [Escherichia coli K-12],3E1F_2 Chain 2, Beta-galactosidase [Escherichia coli K-12],3E1F_3 Chain 3, Beta-galactosidase [Escherichia coli K-12],3E1F_4 Chain 4, Beta-galactosidase [Escherichia coli K-12] |
| 3DYO_A | 8.66e-18 | 106 | 483 | 123 | 499 | ChainA, Beta-galactosidase [Escherichia coli K-12],3DYO_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYO_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYO_D Chain D, Beta-galactosidase [Escherichia coli K-12],3DYP_A Chain A, Beta-galactosidase [Escherichia coli K-12],3DYP_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYP_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYP_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
| 1F4A_A | 1.13e-17 | 106 | 483 | 121 | 497 | E.COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_B E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_C E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_D E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4H_A E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_B E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_C E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_D E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli] |
| 3MUY_1 | 1.14e-17 | 106 | 483 | 123 | 499 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
| 7BRS_A | 1.14e-17 | 106 | 483 | 125 | 501 | ChainA, Beta-galactosidase [Escherichia coli K-12],7BRS_B Chain B, Beta-galactosidase [Escherichia coli K-12],7BRS_C Chain C, Beta-galactosidase [Escherichia coli K-12],7BRS_D Chain D, Beta-galactosidase [Escherichia coli K-12],7BTK_A Chain A, Beta-galactosidase [Escherichia coli K-12],7BTK_B Chain B, Beta-galactosidase [Escherichia coli K-12],7BTK_C Chain C, Beta-galactosidase [Escherichia coli K-12],7BTK_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1LIM9 | 7.09e-18 | 117 | 483 | 135 | 500 | Beta-galactosidase OS=Escherichia coli (strain SMS-3-5 / SECEC) OX=439855 GN=lacZ PE=3 SV=1 |
| B7N8Q1 | 9.30e-18 | 106 | 483 | 124 | 500 | Beta-galactosidase OS=Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC) OX=585056 GN=lacZ PE=3 SV=1 |
| A8AKB8 | 1.22e-17 | 106 | 483 | 124 | 500 | Beta-galactosidase OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=lacZ PE=3 SV=1 |
| Q3Z583 | 1.22e-17 | 106 | 483 | 124 | 500 | Beta-galactosidase OS=Shigella sonnei (strain Ss046) OX=300269 GN=lacZ PE=3 SV=1 |
| A7MN76 | 1.23e-17 | 106 | 542 | 134 | 554 | Beta-galactosidase OS=Cronobacter sakazakii (strain ATCC BAA-894) OX=290339 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
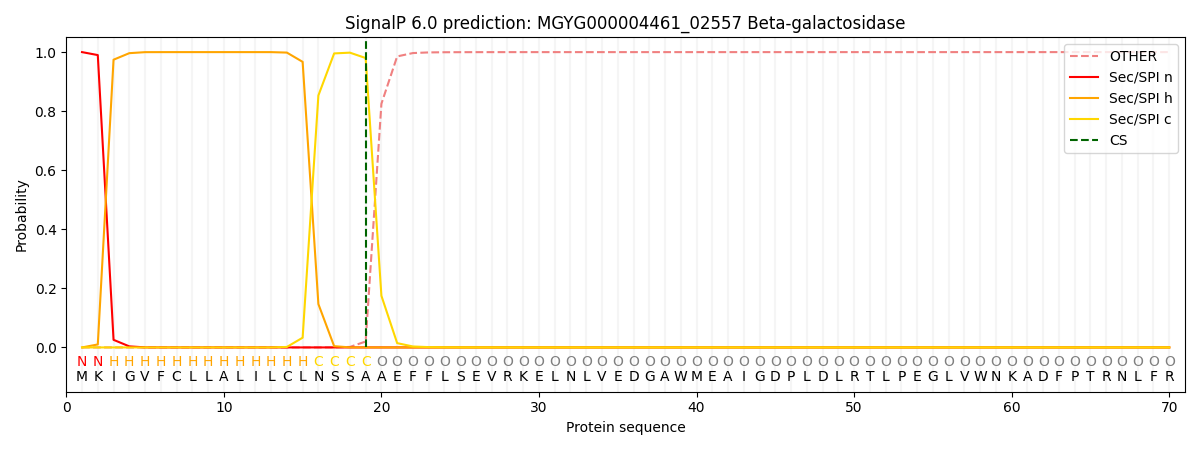
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000254 | 0.999144 | 0.000153 | 0.000152 | 0.000130 | 0.000134 |
