You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004461_02683
You are here: Home > Sequence: MGYG000004461_02683
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
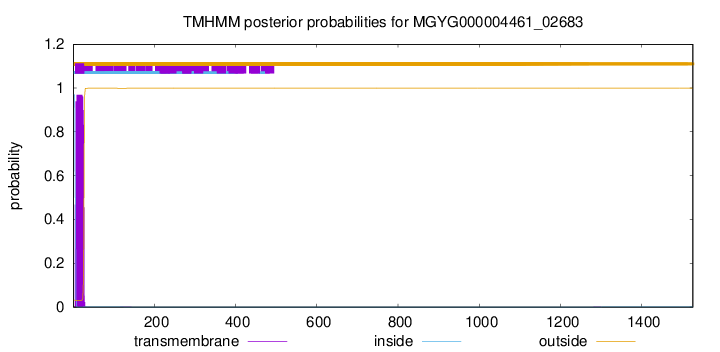
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; ; | |||||||||||
| CAZyme ID | MGYG000004461_02683 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26319; End: 30899 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 29 | 740 | 4.8e-59 | 0.7486702127659575 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.67e-26 | 105 | 464 | 66 | 430 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 5.43e-24 | 29 | 463 | 10 | 445 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 6.12e-15 | 106 | 502 | 113 | 507 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 2.94e-14 | 106 | 433 | 124 | 462 | beta-galactosidase. |
| pfam00703 | Glyco_hydro_2 | 1.64e-10 | 208 | 317 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHF94339.1 | 5.39e-309 | 27 | 1524 | 45 | 1608 |
| QUH31336.1 | 8.44e-44 | 112 | 1044 | 67 | 1035 |
| AVM46955.1 | 2.54e-38 | 102 | 1316 | 126 | 1176 |
| AVM45809.1 | 3.45e-34 | 106 | 1469 | 129 | 1339 |
| CQR58310.1 | 1.54e-30 | 29 | 1112 | 2 | 1169 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T98_A | 9.88e-23 | 103 | 557 | 85 | 530 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 6XXW_A | 2.33e-16 | 105 | 576 | 75 | 575 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 3IAP_A | 3.39e-15 | 33 | 562 | 53 | 575 | ChainA, Beta-galactosidase [Escherichia coli K-12],3IAP_B Chain B, Beta-galactosidase [Escherichia coli K-12],3IAP_C Chain C, Beta-galactosidase [Escherichia coli K-12],3IAP_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
| 3DYM_A | 3.39e-15 | 33 | 562 | 53 | 575 | ChainA, Beta-galactosidase [Escherichia coli K-12],3DYM_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYM_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYM_D Chain D, Beta-galactosidase [Escherichia coli K-12],3E1F_1 Chain 1, Beta-galactosidase [Escherichia coli K-12],3E1F_2 Chain 2, Beta-galactosidase [Escherichia coli K-12],3E1F_3 Chain 3, Beta-galactosidase [Escherichia coli K-12],3E1F_4 Chain 4, Beta-galactosidase [Escherichia coli K-12] |
| 3J7H_A | 4.45e-15 | 33 | 562 | 54 | 576 | Structureof beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],3J7H_B Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],3J7H_C Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],3J7H_D Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],4CKD_A Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],4CKD_B Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],4CKD_C Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],4CKD_D Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],6DRV_A Beta-galactosidase [Escherichia coli K-12],6DRV_B Beta-galactosidase [Escherichia coli K-12],6DRV_C Beta-galactosidase [Escherichia coli K-12],6DRV_D Beta-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 1.10e-20 | 104 | 436 | 105 | 433 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| Q03WL0 | 3.15e-18 | 30 | 571 | 59 | 576 | Beta-galactosidase OS=Leuconostoc mesenteroides subsp. mesenteroides (strain ATCC 8293 / DSM 20343 / BCRC 11652 / CCM 1803 / JCM 6124 / NCDO 523 / NBRC 100496 / NCIMB 8023 / NCTC 12954 / NRRL B-1118 / 37Y) OX=203120 GN=lacZ PE=3 SV=1 |
| Q04F24 | 3.18e-16 | 56 | 532 | 68 | 534 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
| T2KM09 | 4.18e-16 | 105 | 517 | 109 | 508 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| Q3Z583 | 9.38e-16 | 33 | 562 | 54 | 576 | Beta-galactosidase OS=Shigella sonnei (strain Ss046) OX=300269 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
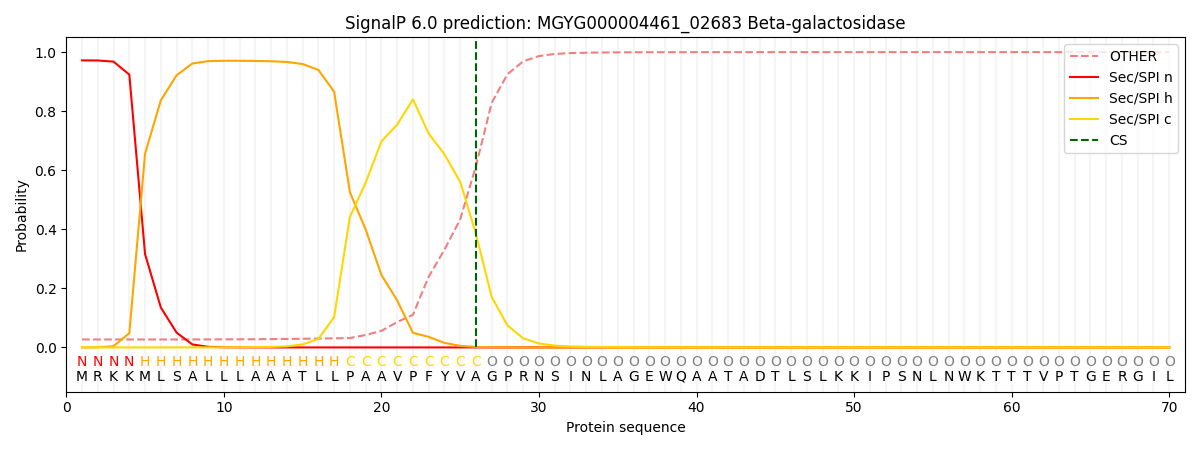
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.030216 | 0.967146 | 0.001631 | 0.000401 | 0.000296 | 0.000296 |