You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004471_01819
You are here: Home > Sequence: MGYG000004471_01819
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
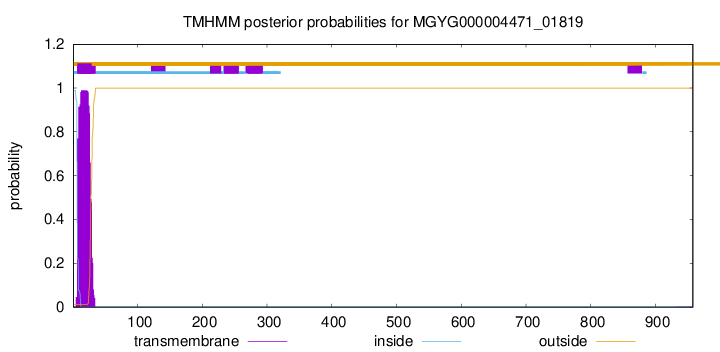
TMHMM annotations
Basic Information help
| Species | UBA1394 sp900554975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1394; UBA1394 sp900554975 | |||||||||||
| CAZyme ID | MGYG000004471_01819 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3487; End: 6363 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 52 | 237 | 5.8e-73 | 0.9943502824858758 |
| CE1 | 533 | 768 | 2.3e-35 | 0.9647577092511013 |
| CBM22 | 264 | 391 | 8.1e-33 | 0.9694656488549618 |
| CBM22 | 789 | 916 | 8.8e-31 | 0.9618320610687023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 1.25e-75 | 52 | 235 | 1 | 175 | Glycosyl hydrolases family 11. |
| pfam02018 | CBM_4_9 | 2.60e-23 | 261 | 395 | 1 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| COG2382 | Fes | 1.05e-22 | 520 | 772 | 65 | 298 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| cd14256 | Dockerin_I | 6.72e-18 | 420 | 472 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam02018 | CBM_4_9 | 2.82e-17 | 788 | 920 | 4 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAB93667.1 | 2.07e-201 | 46 | 771 | 32 | 792 |
| AAR39816.1 | 8.97e-193 | 40 | 742 | 28 | 782 |
| CAA90271.1 | 8.97e-193 | 40 | 742 | 28 | 782 |
| CBL18305.1 | 8.15e-192 | 6 | 958 | 7 | 668 |
| CBL17231.1 | 1.43e-129 | 208 | 772 | 507 | 1074 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7AYL_A | 7.23e-85 | 49 | 245 | 38 | 231 | Crystalstructure of the GH11 domain of a multidomain xylanase from the hindgut metagenome of Trinervitermes trinervoides [uncultured bacterium],7AYL_B Crystal structure of the GH11 domain of a multidomain xylanase from the hindgut metagenome of Trinervitermes trinervoides [uncultured bacterium] |
| 2F6B_A | 7.02e-83 | 45 | 245 | 5 | 203 | Structuraland active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus],2F6B_B Structural and active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus] |
| 1H4G_A | 1.41e-82 | 45 | 245 | 5 | 203 | Oligosaccharide-bindingto family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1H4G_B Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1QH6_A CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens],1QH6_B CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens],1QH7_A CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens],1QH7_B CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens] |
| 6KKA_A | 1.92e-82 | 49 | 246 | 8 | 203 | XylanaseJ mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],6KKA_B Xylanase J mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 1IGO_A | 2.54e-82 | 49 | 246 | 10 | 203 | Family11 xylanase [Bacillus subtilis],1IGO_B Family 11 xylanase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q53317 | 2.29e-102 | 41 | 440 | 30 | 458 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
| P17137 | 1.71e-83 | 49 | 244 | 68 | 260 | Endo-1,4-beta-xylanase OS=Clostridium saccharobutylicum OX=169679 GN=xynB PE=3 SV=1 |
| Q8GJ44 | 8.11e-83 | 1 | 259 | 1 | 247 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P33558 | 1.19e-79 | 1 | 259 | 1 | 248 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P83513 | 2.27e-79 | 31 | 245 | 9 | 217 | Bifunctional xylanase/deacetylase OS=Pseudobutyrivibrio xylanivorans OX=185007 GN=xyn11A PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000236 | 0.999103 | 0.000203 | 0.000171 | 0.000156 | 0.000142 |