You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004490_00210
You are here: Home > Sequence: MGYG000004490_00210
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muribaculum sp003150235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp003150235 | |||||||||||
| CAZyme ID | MGYG000004490_00210 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66709; End: 69075 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 498 | 761 | 4.1e-57 | 0.6897689768976898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.79e-47 | 491 | 758 | 45 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.30e-40 | 494 | 764 | 113 | 343 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 5.92e-40 | 494 | 760 | 90 | 310 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 2.28e-10 | 76 | 138 | 22 | 85 | Glycosyl hydrolase family 10. |
| pfam02018 | CBM_4_9 | 1.14e-08 | 164 | 281 | 2 | 123 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP72441.1 | 0.0 | 1 | 784 | 1 | 791 |
| QUT92890.1 | 1.30e-293 | 1 | 787 | 1 | 769 |
| ALJ61540.1 | 1.05e-292 | 1 | 787 | 1 | 769 |
| EDV05054.1 | 3.98e-262 | 1 | 787 | 1 | 778 |
| QDO69424.1 | 2.27e-261 | 1 | 787 | 1 | 778 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2F8Q_A | 8.81e-26 | 478 | 760 | 107 | 353 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 2FGL_A | 8.96e-26 | 478 | 760 | 108 | 354 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2FGL_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 4QDM_A | 9.11e-26 | 478 | 760 | 109 | 355 | Crystalstructure of N-terminal mutant (V1L) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],4QDM_B Crystal structure of N-terminal mutant (V1L) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 5EB8_A | 9.11e-26 | 478 | 760 | 109 | 355 | Crystalstructure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5EB8_B Crystal structure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 4QCF_A | 9.11e-26 | 478 | 760 | 109 | 355 | Crystalstructure of N-terminal mutant (V1A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P36917 | 1.46e-26 | 103 | 760 | 1 | 674 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P07528 | 1.26e-24 | 498 | 760 | 162 | 396 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| P38535 | 9.61e-23 | 557 | 760 | 343 | 526 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P14768 | 3.16e-22 | 512 | 764 | 372 | 610 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| Q59675 | 5.16e-21 | 507 | 770 | 359 | 605 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
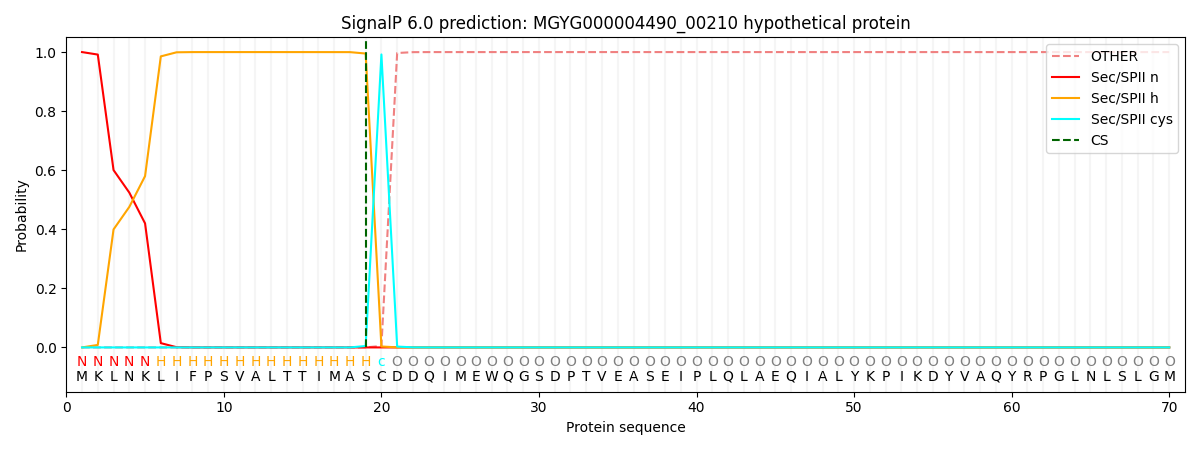
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000061 | 0.000000 | 0.000000 | 0.000000 |
