You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004490_01682
You are here: Home > Sequence: MGYG000004490_01682
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muribaculum sp003150235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp003150235 | |||||||||||
| CAZyme ID | MGYG000004490_01682 | |||||||||||
| CAZy Family | GH53 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 74388; End: 75461 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 51 | 335 | 3.9e-87 | 0.9035087719298246 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07745 | Glyco_hydro_53 | 1.91e-90 | 50 | 354 | 1 | 333 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| COG3867 | GanB | 3.07e-78 | 12 | 354 | 5 | 391 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| cd19608 | GH113_mannanase-like | 6.90e-06 | 64 | 350 | 10 | 304 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
| cd11330 | AmyAc_OligoGlu | 2.52e-04 | 87 | 151 | 41 | 148 | Alpha amylase catalytic domain found in oligo-1,6-glucosidase (also called isomaltase; sucrase-isomaltase; alpha-limit dextrinase) and related proteins. Oligo-1,6-glucosidase (EC 3.2.1.10) hydrolyzes the alpha-1,6-glucosidic linkage of isomalto-oligosaccharides, pannose, and dextran. Unlike alpha-1,4-glucosidases (EC 3.2.1.20), it fails to hydrolyze the alpha-1,4-glucosidic bonds of maltosaccharides. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXV50242.1 | 1.36e-113 | 21 | 356 | 21 | 349 |
| QUB71503.1 | 2.65e-113 | 21 | 356 | 20 | 348 |
| QUB93862.1 | 3.88e-113 | 21 | 356 | 21 | 349 |
| QUB91945.1 | 1.10e-112 | 21 | 356 | 21 | 349 |
| QUI93100.1 | 1.57e-112 | 21 | 356 | 21 | 349 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GPA_A | 4.66e-107 | 43 | 354 | 1 | 313 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482],6GPA_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482] |
| 6GP5_A | 1.15e-106 | 23 | 354 | 21 | 349 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],6GP5_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4BF7_A | 1.19e-38 | 51 | 342 | 22 | 329 | Emericillanidulans endo-beta-1,4-galactanase [Aspergillus nidulans] |
| 7OSK_A | 3.48e-37 | 6 | 299 | 14 | 298 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 1FHL_A | 3.24e-36 | 51 | 342 | 5 | 313 | CrystalStructure Of Beta-1,4-galactanase From Aspergillus Aculeatus At 293k [Aspergillus aculeatus],1FOB_A Crystal Structure Of Beta-1,4-galactanase From Aspergillus Aculeatus At 100k [Aspergillus aculeatus],6Q3R_A ASPERGILLUS ACULEATUS GALACTANASE [Aspergillus aculeatus],6Q3R_B ASPERGILLUS ACULEATUS GALACTANASE [Aspergillus aculeatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48843 | 6.58e-45 | 48 | 322 | 5 | 282 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| Q5B153 | 6.23e-38 | 51 | 342 | 22 | 329 | Arabinogalactan endo-beta-1,4-galactanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=galA PE=1 SV=2 |
| Q0CTQ7 | 2.31e-37 | 41 | 342 | 9 | 328 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=galA PE=3 SV=1 |
| Q9Y7F8 | 3.29e-37 | 51 | 342 | 21 | 329 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus tubingensis OX=5068 GN=galA PE=2 SV=1 |
| O07013 | 1.02e-35 | 49 | 328 | 53 | 351 | Endo-beta-1,4-galactanase OS=Bacillus subtilis (strain 168) OX=224308 GN=ganB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
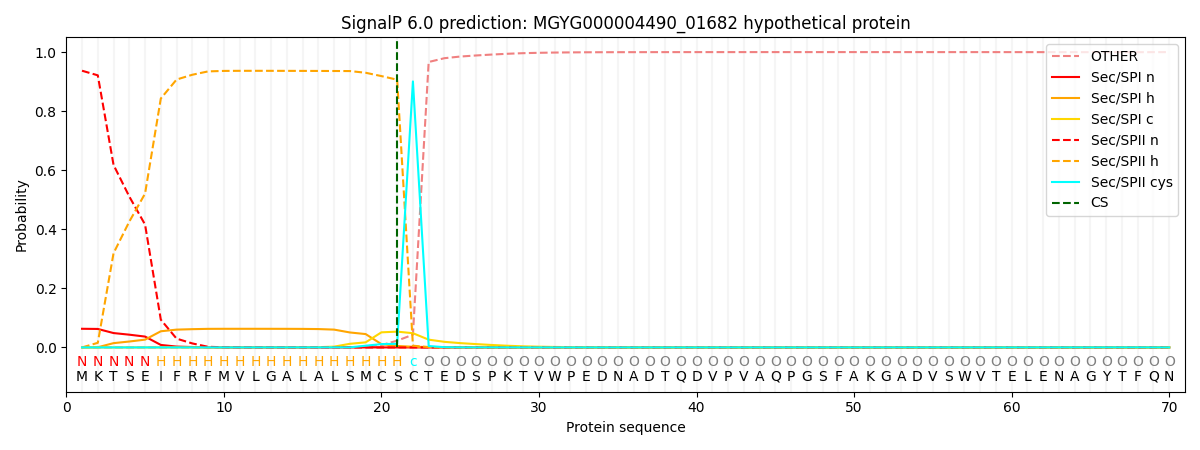
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000081 | 0.061329 | 0.938539 | 0.000028 | 0.000038 | 0.000026 |
