You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004491_01956
You are here: Home > Sequence: MGYG000004491_01956
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
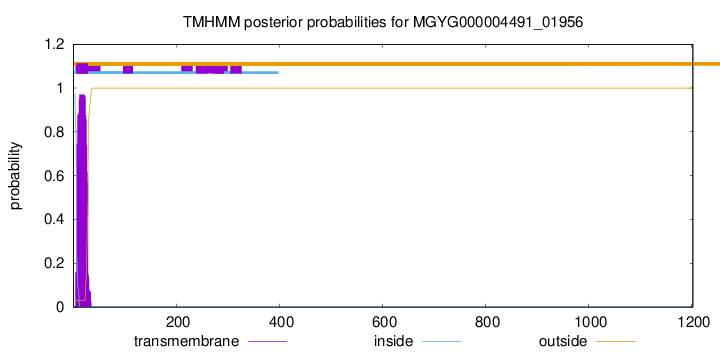
TMHMM annotations
Basic Information help
| Species | Prevotella sp900548745 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900548745 | |||||||||||
| CAZyme ID | MGYG000004491_01956 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 483; End: 4091 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 641 | 933 | 9.7e-48 | 0.9409722222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01095 | Pectinesterase | 2.30e-24 | 676 | 935 | 37 | 297 | Pectinesterase. |
| COG4677 | PemB | 2.77e-18 | 711 | 855 | 175 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02713 | PLN02713 | 4.73e-17 | 737 | 939 | 346 | 552 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02916 | PLN02916 | 1.73e-16 | 629 | 939 | 185 | 490 | pectinesterase family protein |
| PLN02314 | PLN02314 | 3.49e-16 | 676 | 937 | 315 | 573 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB37302.1 | 1.00e-172 | 27 | 1064 | 30 | 1141 |
| ANU64595.1 | 1.00e-172 | 27 | 1064 | 30 | 1141 |
| QQR08039.1 | 1.00e-172 | 27 | 1064 | 30 | 1141 |
| QUT74167.1 | 1.53e-58 | 429 | 1015 | 795 | 1382 |
| QCD38476.1 | 8.60e-58 | 492 | 1010 | 901 | 1416 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
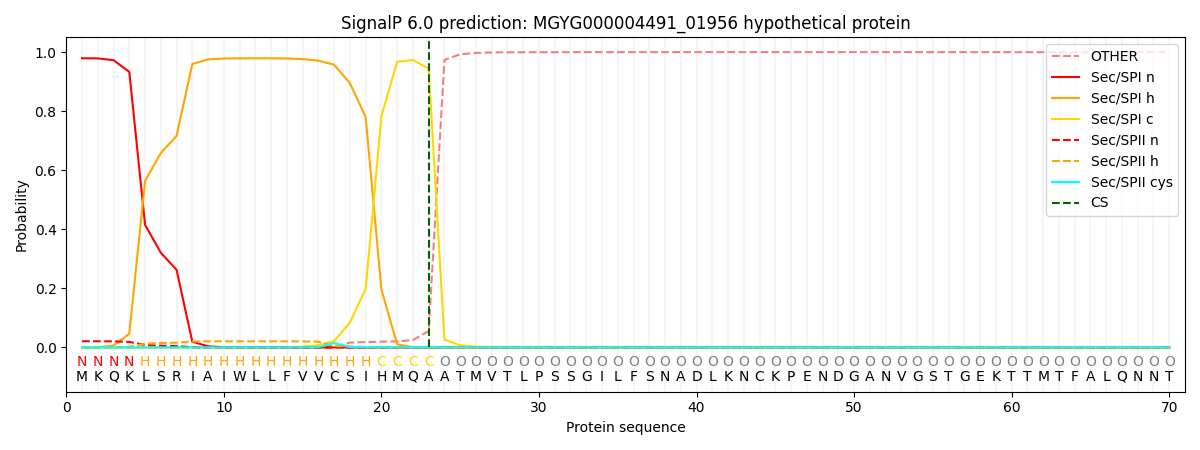
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000686 | 0.975062 | 0.023468 | 0.000270 | 0.000245 | 0.000234 |