You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004493_00023
You are here: Home > Sequence: MGYG000004493_00023
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Spirochaetota; Spirochaetia; Treponematales; Treponemataceae; Treponema_D; | |||||||||||
| CAZyme ID | MGYG000004493_00023 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19692; End: 20783 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 101 | 314 | 1.4e-36 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 5.42e-44 | 35 | 349 | 4 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 8.58e-42 | 35 | 359 | 5 | 319 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 1.57e-23 | 66 | 292 | 27 | 256 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 2.44e-05 | 199 | 359 | 204 | 360 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEB13960.1 | 2.87e-220 | 1 | 362 | 1 | 362 |
| QQA00274.1 | 1.20e-112 | 30 | 359 | 33 | 364 |
| QSI02896.1 | 5.11e-97 | 30 | 362 | 56 | 395 |
| QTQ11429.1 | 2.87e-92 | 30 | 358 | 64 | 409 |
| QTQ16630.1 | 2.87e-92 | 30 | 358 | 64 | 409 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VI6_A | 1.09e-23 | 65 | 315 | 26 | 276 | ChainA, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI6_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_A Chain A, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835] |
| 5BZA_A | 1.16e-21 | 55 | 336 | 15 | 297 | Crystalstructure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5BZA_B Crystal structure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5BZA_C Crystal structure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5BZA_D Crystal structure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_A Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_B Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_C Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_D Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana] |
| 6GFV_A | 6.37e-21 | 99 | 314 | 72 | 298 | Mtuberculosis LpqI [Mycobacterium tuberculosis H37Rv],6GFV_B M tuberculosis LpqI [Mycobacterium tuberculosis H37Rv] |
| 5G1M_A | 7.08e-21 | 66 | 290 | 48 | 268 | Crystalstructure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G1M_B Crystal structure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G2M_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G2M_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G3R_A Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G3R_B Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G5K_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5G5K_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
| 3WO8_A | 3.37e-20 | 65 | 336 | 40 | 308 | Crystalstructure of the beta-N-acetylglucosaminidase from Thermotoga maritima [Thermotoga maritima MSB8],3WO8_B Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P44955 | 8.40e-25 | 42 | 340 | 5 | 304 | Beta-hexosaminidase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=nagZ PE=3 SV=1 |
| P48823 | 5.50e-24 | 66 | 315 | 65 | 335 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| Q0AF74 | 7.49e-24 | 66 | 292 | 29 | 258 | Beta-hexosaminidase OS=Nitrosomonas eutropha (strain DSM 101675 / C91 / Nm57) OX=335283 GN=nagZ PE=3 SV=1 |
| Q4QLU8 | 1.07e-23 | 42 | 340 | 5 | 304 | Beta-hexosaminidase OS=Haemophilus influenzae (strain 86-028NP) OX=281310 GN=nagZ PE=3 SV=1 |
| A5UDA9 | 5.17e-23 | 42 | 340 | 5 | 304 | Beta-hexosaminidase OS=Haemophilus influenzae (strain PittEE) OX=374930 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
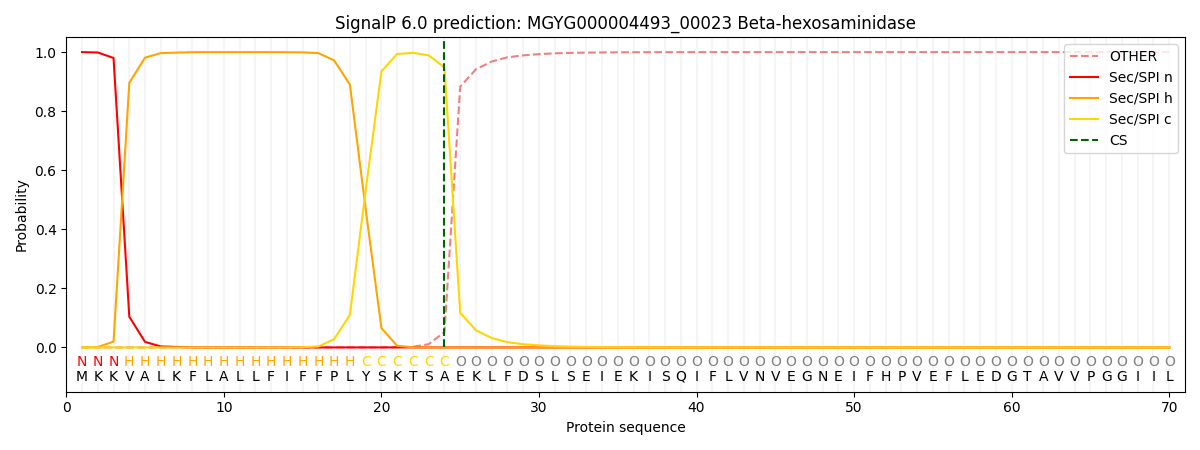
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000523 | 0.998451 | 0.000465 | 0.000186 | 0.000187 | 0.000169 |
