You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004493_01583
You are here: Home > Sequence: MGYG000004493_01583
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Spirochaetota; Spirochaetia; Treponematales; Treponemataceae; Treponema_D; | |||||||||||
| CAZyme ID | MGYG000004493_01583 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48589; End: 50718 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 61 | 370 | 4.2e-96 | 0.9896193771626297 |
| CBM27 | 537 | 705 | 1.7e-21 | 0.9880952380952381 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 2.84e-48 | 38 | 708 | 4 | 583 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam09212 | CBM27 | 1.21e-23 | 527 | 708 | 1 | 173 | Carbohydrate binding module 27. Members of this family are carbohydrate binding modules that bind to beta-1, 4-manno-oligosaccharides, carob galactomannan, and konjac glucomannan, but not to cellulose (insoluble and soluble) or soluble birchwood xylan. They adopt a beta sandwich structure comprising 13 beta strands with a single, small alpha-helix and a single metal atom. |
| pfam00150 | Cellulase | 1.68e-10 | 41 | 354 | 4 | 252 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEE17337.1 | 6.94e-296 | 14 | 709 | 29 | 726 |
| QCK83069.1 | 5.98e-159 | 30 | 708 | 47 | 692 |
| AAC71692.1 | 1.23e-158 | 30 | 708 | 47 | 693 |
| QXJ38243.1 | 3.36e-158 | 30 | 708 | 47 | 692 |
| ACM23521.1 | 1.24e-137 | 30 | 708 | 23 | 664 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6TN6_A | 1.65e-132 | 39 | 709 | 7 | 641 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 3PZ9_A | 5.83e-114 | 33 | 413 | 15 | 383 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 4QP0_A | 2.39e-69 | 27 | 369 | 1 | 324 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 4AWE_A | 7.95e-58 | 28 | 369 | 4 | 344 | TheCrystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase [Neurospora sitophila] |
| 3ZIZ_A | 1.13e-51 | 17 | 409 | 2 | 350 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O48540 | 3.00e-59 | 22 | 371 | 21 | 356 | Mannan endo-1,4-beta-mannosidase 1 OS=Solanum lycopersicum OX=4081 GN=MAN1 PE=1 SV=2 |
| Q9FUQ6 | 1.58e-54 | 16 | 371 | 14 | 358 | Mannan endo-1,4-beta-mannosidase 3 OS=Solanum lycopersicum OX=4081 GN=MAN3 PE=3 SV=1 |
| Q4WBS1 | 1.40e-53 | 25 | 369 | 92 | 402 | Mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=manF PE=1 SV=2 |
| B0Y9E7 | 1.40e-53 | 25 | 369 | 92 | 402 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
| Q5AZ53 | 1.35e-51 | 19 | 410 | 11 | 389 | Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
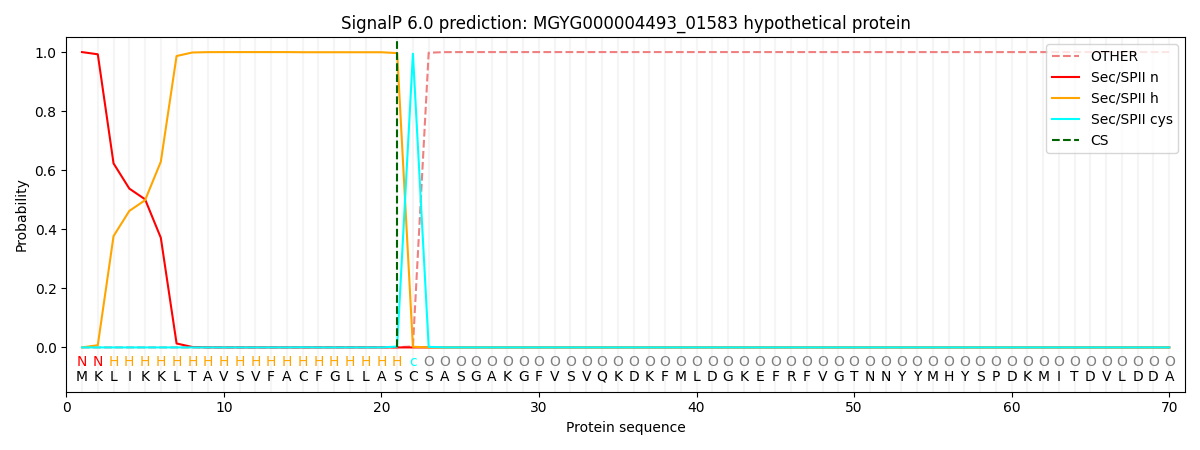
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000070 | 0.000000 | 0.000000 | 0.000000 |
