You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004495_01308
You are here: Home > Sequence: MGYG000004495_01308
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004495_01308 | |||||||||||
| CAZy Family | GH66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18446; End: 20212 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH66 | 45 | 587 | 1.6e-182 | 0.9946043165467626 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13199 | Glyco_hydro_66 | 0.0 | 48 | 585 | 1 | 557 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
| cd14745 | GH66 | 7.40e-165 | 133 | 459 | 1 | 331 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85291.1 | 0.0 | 1 | 588 | 1 | 588 |
| QDO70249.1 | 1.36e-246 | 1 | 587 | 1 | 596 |
| QUT91748.1 | 2.23e-245 | 1 | 587 | 1 | 596 |
| ALJ62514.1 | 1.28e-244 | 1 | 587 | 1 | 596 |
| QUT36786.1 | 3.38e-236 | 1 | 587 | 1 | 601 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AXG_A | 1.80e-111 | 45 | 588 | 46 | 610 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXG_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223] |
| 5AXH_A | 1.97e-110 | 45 | 588 | 46 | 610 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXH_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose [Thermoanaerobacter pseudethanolicus ATCC 33223] |
| 3WNK_A | 5.62e-70 | 48 | 584 | 32 | 719 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNP_A | 4.61e-69 | 48 | 584 | 13 | 700 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNP_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNL_A | 6.40e-69 | 48 | 584 | 13 | 700 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94286 | 3.16e-68 | 48 | 588 | 49 | 740 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
| P70873 | 1.05e-67 | 48 | 588 | 41 | 732 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 GN=cit PE=3 SV=1 |
| P39653 | 6.74e-33 | 95 | 588 | 231 | 802 | Dextranase OS=Streptococcus downei OX=1317 GN=dex PE=1 SV=1 |
| Q54443 | 6.44e-29 | 44 | 588 | 106 | 734 | Dextranase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=dexA PE=1 SV=2 |
| Q59979 | 1.90e-26 | 99 | 588 | 42 | 599 | Dextranase OS=Streptococcus salivarius OX=1304 GN=dex PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
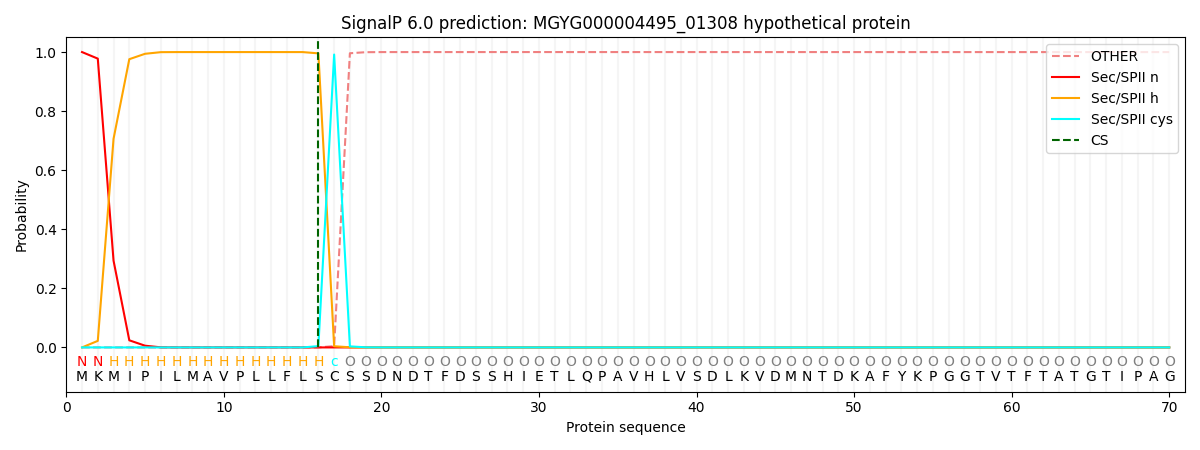
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000088 | 0.000000 | 0.000000 | 0.000000 |
