You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004501_00305
You are here: Home > Sequence: MGYG000004501_00305
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760665 | |||||||||||
| CAZyme ID | MGYG000004501_00305 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4728; End: 6329 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 82 | 483 | 1.6e-39 | 0.9636963696369637 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.01e-26 | 147 | 483 | 3 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.19e-25 | 147 | 483 | 46 | 308 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.81e-19 | 147 | 489 | 69 | 343 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM47074.1 | 3.24e-176 | 21 | 533 | 3 | 496 |
| QQZ02681.1 | 1.09e-175 | 30 | 532 | 19 | 494 |
| QGA28189.1 | 9.41e-174 | 26 | 524 | 18 | 495 |
| AHF92621.1 | 6.37e-167 | 30 | 525 | 12 | 483 |
| AWI10666.1 | 6.35e-159 | 36 | 533 | 1 | 480 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VBR_A | 4.36e-14 | 142 | 490 | 48 | 323 | Crystalstructure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
| 3NIY_A | 1.58e-13 | 142 | 490 | 64 | 339 | Crystalstructure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
| 3W24_A | 2.84e-11 | 165 | 486 | 69 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 4W8L_A | 4.21e-11 | 154 | 483 | 56 | 341 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 7NL2_A | 5.61e-11 | 155 | 489 | 65 | 343 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q60041 | 2.20e-11 | 143 | 492 | 68 | 344 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
| O69230 | 3.82e-11 | 154 | 503 | 422 | 734 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| Q12603 | 5.45e-11 | 153 | 483 | 70 | 349 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
| Q4P902 | 2.20e-09 | 95 | 488 | 33 | 340 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| O59859 | 2.64e-09 | 147 | 483 | 77 | 323 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
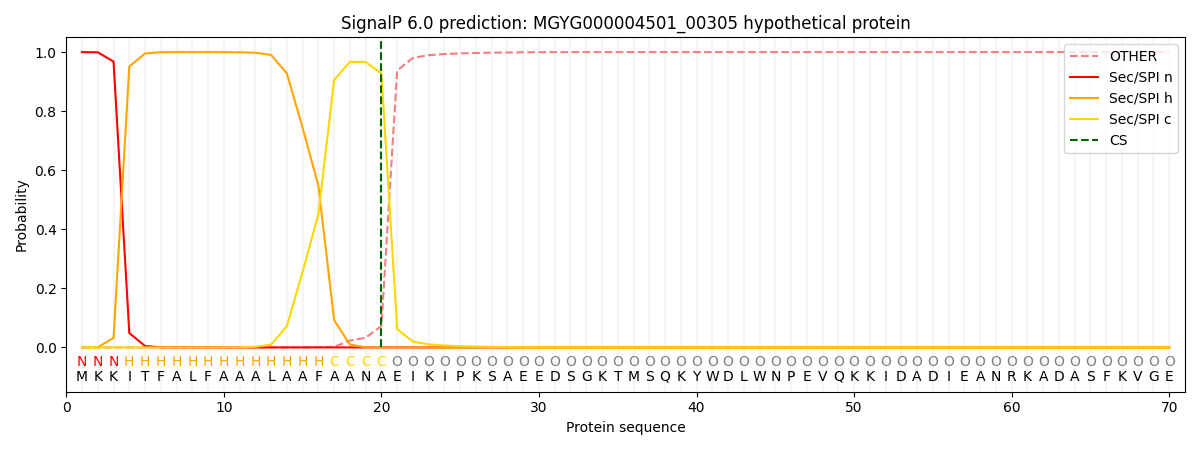
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.999058 | 0.000166 | 0.000169 | 0.000157 | 0.000143 |
