You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004513_00407
You are here: Home > Sequence: MGYG000004513_00407
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900550905 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900550905 | |||||||||||
| CAZyme ID | MGYG000004513_00407 | |||||||||||
| CAZy Family | PL21 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20405; End: 23560 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL21 | 692 | 758 | 7.3e-26 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16286 | SPM-1-like_MBL-B1-B2-like | 0.009 | 131 | 200 | 94 | 155 | Pseudomonas areoginosa SPM-1 and related metallo-beta-lactamases, subclasses B1 and B2 like; MBL-fold metallo-hydrolase domain. SPM-1 was first identified in a Pseudomonas aeruginosa strain from a paediatric leukaemia patient and is a major clinical problem. MBLs (class B of the Ambler beta-lactamase classification) have been divided into three subclasses B1, B2 and B3, based on sequence/structural relationships and substrates, with the B1 and B2 MBLs are most closely related to each other. SPM-1 appears to be a hybrid B1/B2 MBL. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWO00644.1 | 6.19e-178 | 199 | 1048 | 51 | 906 |
| ADY51260.1 | 3.87e-173 | 197 | 1021 | 48 | 877 |
| SCD19150.1 | 7.00e-160 | 183 | 1015 | 35 | 880 |
| QGA27893.1 | 1.29e-159 | 191 | 1015 | 1 | 830 |
| AEW00072.1 | 1.09e-158 | 274 | 1047 | 111 | 882 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2FUQ_A | 9.03e-101 | 338 | 1015 | 18 | 714 | ChainA, heparinase II protein [Pedobacter heparinus],2FUQ_B Chain B, heparinase II protein [Pedobacter heparinus] |
| 3E80_A | 9.47e-101 | 338 | 1015 | 20 | 716 | Structureof Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus],3E80_B Structure of Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus],3E80_C Structure of Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus] |
| 3E7J_A | 4.73e-98 | 338 | 1015 | 20 | 716 | ChainA, Heparinase II protein [Pedobacter heparinus],3E7J_B Chain B, Heparinase II protein [Pedobacter heparinus] |
| 2FUT_A | 4.23e-94 | 338 | 1015 | 19 | 715 | ChainA, heparinase II protein [Pedobacter heparinus],2FUT_B Chain B, heparinase II protein [Pedobacter heparinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6XZB6 | 8.90e-100 | 338 | 1015 | 43 | 739 | Heparin and heparin-sulfate lyase OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=hepB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
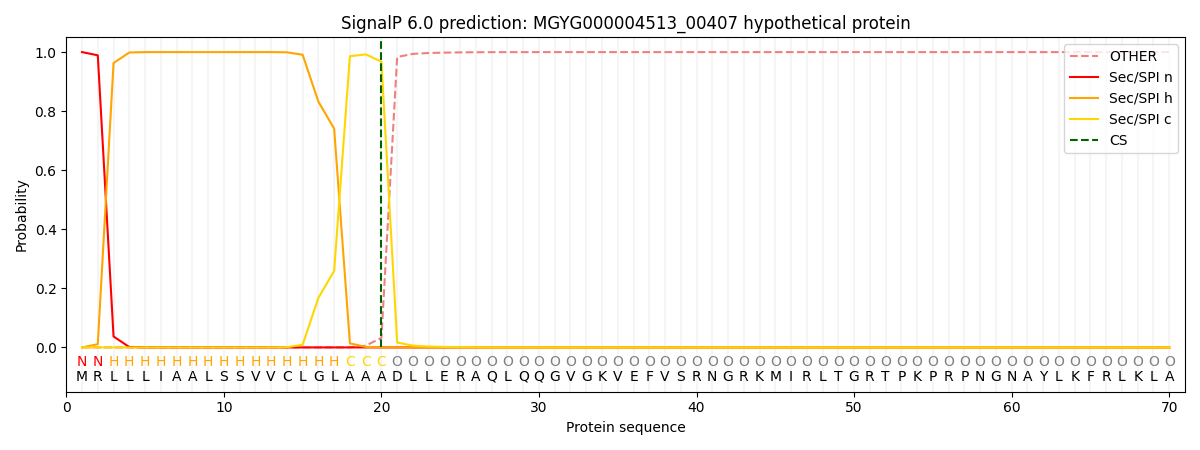
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999170 | 0.000153 | 0.000155 | 0.000148 | 0.000139 |
