You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004513_00497
You are here: Home > Sequence: MGYG000004513_00497
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
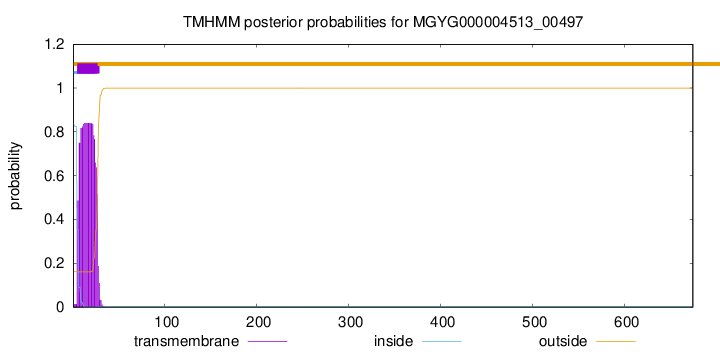
TMHMM annotations
Basic Information help
| Species | Victivallis sp900550905 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900550905 | |||||||||||
| CAZyme ID | MGYG000004513_00497 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6840; End: 8864 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 141 | 445 | 2.4e-21 | 0.5492700729927007 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 1.26e-06 | 551 | 663 | 29 | 123 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 7.40e-06 | 551 | 657 | 52 | 140 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| PLN02218 | PLN02218 | 5.41e-04 | 102 | 284 | 41 | 206 | polygalacturonase ADPG |
| pfam12708 | Pectate_lyase_3 | 0.008 | 141 | 188 | 2 | 48 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK59395.1 | 3.73e-27 | 143 | 633 | 26 | 481 |
| QDU71501.1 | 1.89e-22 | 54 | 628 | 10 | 593 |
| QHI69810.1 | 2.91e-20 | 141 | 617 | 56 | 493 |
| AHF92261.1 | 5.55e-20 | 131 | 660 | 7 | 506 |
| AHF89502.1 | 3.54e-19 | 167 | 655 | 94 | 584 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
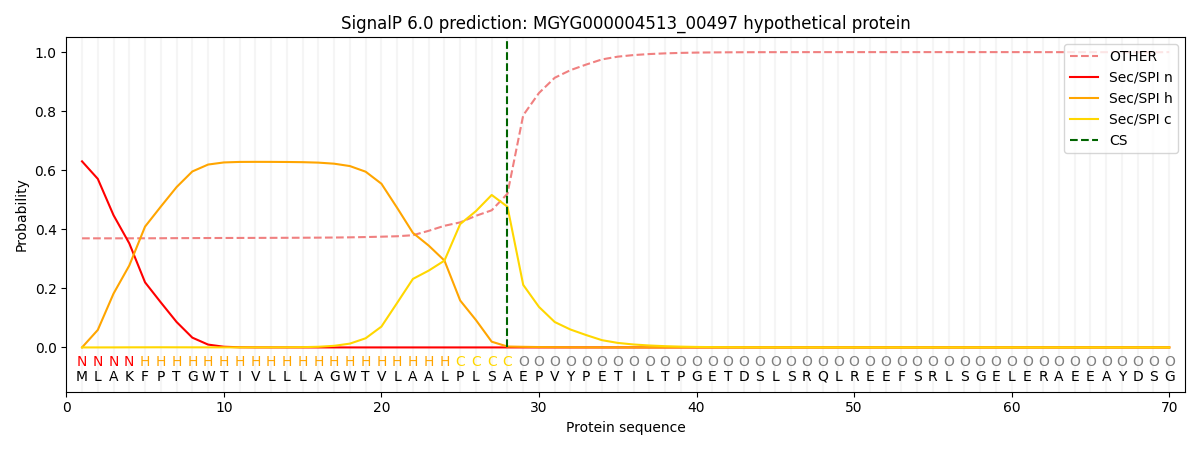
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.377398 | 0.620808 | 0.000935 | 0.000250 | 0.000234 | 0.000367 |