You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004513_02590
You are here: Home > Sequence: MGYG000004513_02590
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900550905 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900550905 | |||||||||||
| CAZyme ID | MGYG000004513_02590 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4195; End: 6201 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 24 | 491 | 2.9e-71 | 0.7702909647779479 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04994 | TfoX_C | 8.61e-04 | 579 | 624 | 34 | 81 | TfoX C-terminal domain. TfoX may play a key role in the development of genetic competence by regulating the expression of late competence-specific genes. This family corresponds to the C-terminal presumed domain of TfoX. The domain is found associated with pfam00383 in Neisseria meningitidis TadA. It is also found as an isolated domain in some proteins suggesting this is an autonomous domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46385.1 | 0.0 | 1 | 668 | 1 | 669 |
| QRO01254.1 | 5.42e-99 | 22 | 501 | 215 | 699 |
| BBI31951.1 | 9.44e-81 | 11 | 495 | 594 | 1100 |
| QBG46742.1 | 8.18e-52 | 24 | 498 | 231 | 749 |
| QZX85855.1 | 2.87e-50 | 24 | 493 | 194 | 699 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XJ9_A | 2.48e-37 | 24 | 491 | 223 | 758 | Structureof PfGH50B [Pseudoalteromonas fuliginea],6XJ9_B Structure of PfGH50B [Pseudoalteromonas fuliginea] |
| 4BQ2_A | 1.34e-36 | 24 | 497 | 204 | 748 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 3.22e-36 | 24 | 497 | 204 | 748 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 5Z6P_A | 6.24e-34 | 24 | 491 | 223 | 758 | Thecrystal structure of an agarase, AgWH50C [Agarivorans gilvus],5Z6P_B The crystal structure of an agarase, AgWH50C [Agarivorans gilvus] |
| 5T3B_A | 1.51e-12 | 67 | 480 | 84 | 461 | ChainA, Glycoside Hydrolase [Phocaeicola plebeius],5T3B_B Chain B, Glycoside Hydrolase [Phocaeicola plebeius] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48840 | 2.15e-36 | 24 | 488 | 432 | 945 | Beta-agarase B OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
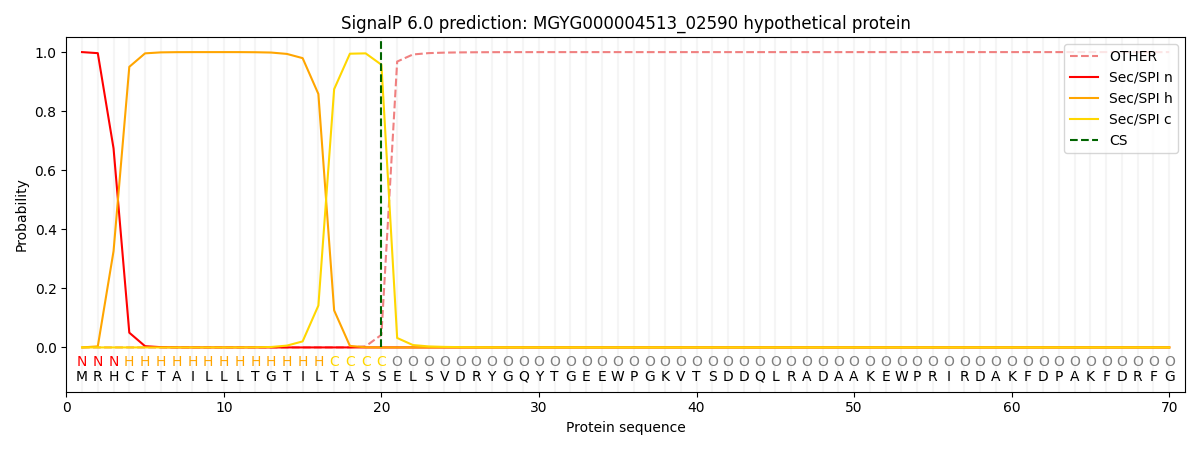
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000290 | 0.999046 | 0.000190 | 0.000164 | 0.000145 | 0.000140 |
