You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004513_02767
You are here: Home > Sequence: MGYG000004513_02767
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900550905 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900550905 | |||||||||||
| CAZyme ID | MGYG000004513_02767 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6353; End: 8218 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 76 | 247 | 1.6e-27 | 0.48259860788863107 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 2.44e-08 | 109 | 247 | 122 | 280 | Glycosyl hydrolases family 39. |
| pfam07745 | Glyco_hydro_53 | 5.16e-07 | 47 | 242 | 30 | 239 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| pfam02449 | Glyco_hydro_42 | 1.25e-06 | 41 | 94 | 10 | 64 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam11790 | Glyco_hydro_cc | 2.64e-05 | 135 | 239 | 70 | 168 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| COG3867 | GanB | 1.84e-04 | 47 | 243 | 69 | 286 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 1.20e-290 | 26 | 617 | 26 | 617 |
| QDU57372.1 | 1.21e-111 | 20 | 618 | 21 | 632 |
| AVM44901.1 | 5.93e-85 | 10 | 586 | 8 | 599 |
| AHF90941.1 | 8.01e-59 | 26 | 604 | 231 | 839 |
| ACZ43011.1 | 1.04e-38 | 27 | 260 | 302 | 546 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 4.48e-14 | 30 | 244 | 107 | 345 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 4ZN2_A | 4.45e-13 | 45 | 255 | 37 | 266 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 4.45e-13 | 45 | 255 | 37 | 266 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
| 1W91_A | 9.20e-07 | 118 | 260 | 138 | 299 | crystalstructure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_B crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_C crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_D crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_E crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_F crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_G crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct],1W91_H crystal structure of 1,4-BETA-D-XYLAN XYLOHYDROLASE solve using anomalous signal from Seleniomethionine [synthetic construct] |
| 2BS9_A | 9.20e-07 | 118 | 260 | 138 | 299 | Nativecrystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_B Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_C Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_D Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_E Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_F Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_G Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus],2BS9_H Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZFM2 | 5.05e-06 | 118 | 260 | 138 | 301 | Beta-xylosidase OS=Geobacillus stearothermophilus OX=1422 GN=xynB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
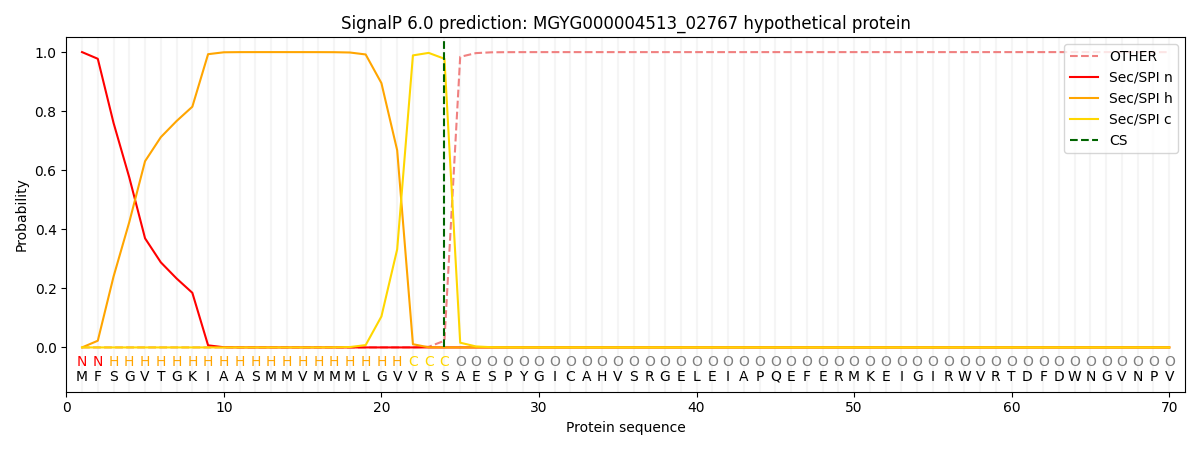
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000371 | 0.998993 | 0.000203 | 0.000146 | 0.000134 | 0.000140 |
