You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004536_00363
You are here: Home > Sequence: MGYG000004536_00363
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900542505 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900542505 | |||||||||||
| CAZyme ID | MGYG000004536_00363 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4693; End: 7098 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06542 | GH18_EndoS-like | 7.88e-50 | 60 | 315 | 1 | 246 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
| pfam16141 | DUF4849 | 9.09e-49 | 25 | 325 | 1 | 316 | Putative glycoside hydrolase Family 18, chitinase_18. This DUF is likely to be a form of glycosyl hydrolase from CAZy family 18, possibly chitinase 18. This would have the EC number of EC:3.2.1.14. |
| pfam08522 | DUF1735 | 2.31e-12 | 336 | 448 | 1 | 111 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam08522 | DUF1735 | 6.46e-09 | 458 | 587 | 1 | 112 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 1.37e-06 | 660 | 794 | 8 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL00953.1 | 1.51e-191 | 1 | 478 | 9 | 481 |
| BBL08878.1 | 1.51e-191 | 1 | 478 | 9 | 481 |
| BBL11670.1 | 1.51e-191 | 1 | 478 | 9 | 481 |
| AFL77898.1 | 1.10e-186 | 1 | 471 | 1 | 468 |
| CBK63302.1 | 3.47e-182 | 1 | 794 | 5 | 770 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q64_A | 1.43e-40 | 3 | 336 | 2 | 363 | BT1044SeMetE190Q [Bacteroides thetaiotaomicron] |
| 6KPL_A | 3.89e-11 | 42 | 330 | 12 | 297 | CrystalStructure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form [Cordyceps militaris CM01],6KPM_A Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose [Cordyceps militaris CM01] |
| 6KPN_A | 4.01e-10 | 42 | 330 | 12 | 297 | CrystalStructure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine [Cordyceps militaris CM01],6KPO_A Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn [Cordyceps militaris CM01] |
| 4NUY_A | 3.87e-06 | 64 | 264 | 19 | 250 | Crystalstructure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes [Streptococcus pyogenes serotype M1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P36912 | 4.97e-11 | 5 | 308 | 6 | 305 | Endo-beta-N-acetylglucosaminidase F2 OS=Elizabethkingia meningoseptica OX=238 GN=endOF2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
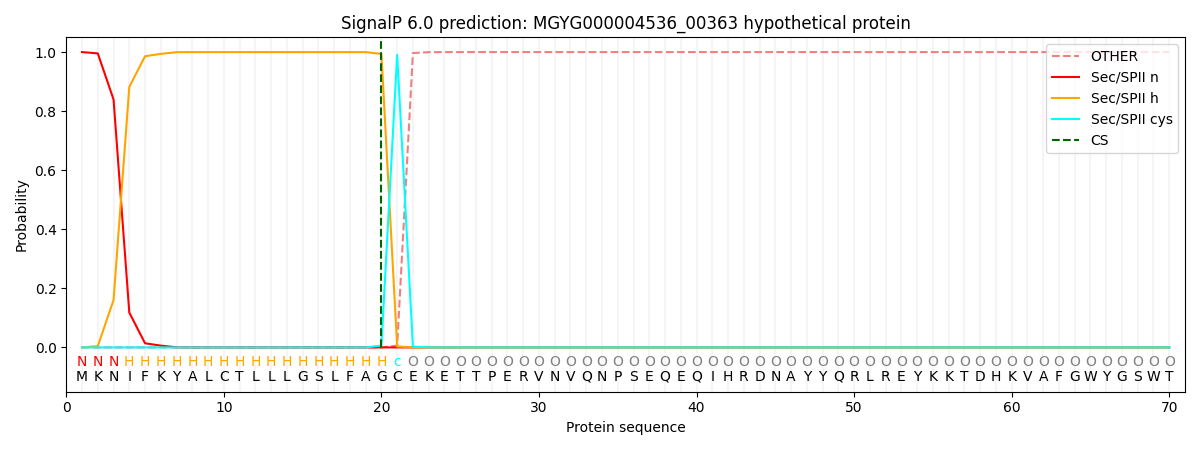
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000064 | 0.000000 | 0.000000 | 0.000000 |
