You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004541_01678
You are here: Home > Sequence: MGYG000004541_01678
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
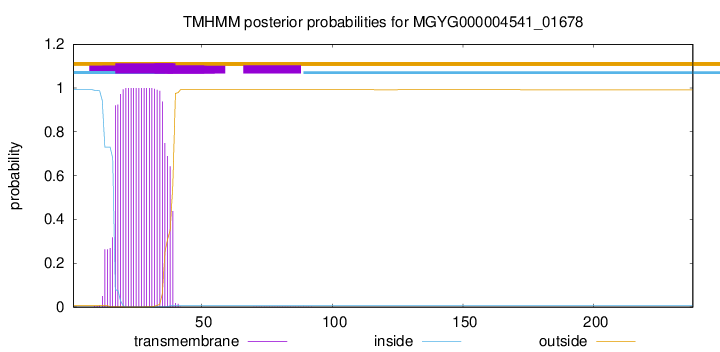
TMHMM annotations
Basic Information help
| Species | Ureibacillus endophyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_A; Planococcaceae; Ureibacillus; Ureibacillus endophyticus | |||||||||||
| CAZyme ID | MGYG000004541_01678 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35127; End: 35843 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 74 | 214 | 2.5e-18 | 0.8814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13399 | Slt35-like | 3.19e-33 | 81 | 209 | 1 | 104 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| COG2951 | MltB | 5.99e-11 | 63 | 189 | 105 | 245 | Membrane-bound lytic murein transglycosylase B [Cell wall/membrane/envelope biogenesis]. |
| pfam13406 | SLT_2 | 3.58e-09 | 69 | 189 | 72 | 205 | Transglycosylase SLT domain. This family is related to the SLT domain pfam01464. |
| pfam01464 | SLT | 4.64e-07 | 76 | 198 | 3 | 93 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd13403 | MLTF-like | 4.77e-06 | 77 | 182 | 4 | 70 | membrane-bound lytic murein transglycosylase F (MLTF) and similar proteins. This subfamily includes membrane-bound lytic murein transglycosylase F (MltF, murein lyase F) that degrades murein glycan strands. It is responsible for catalyzing the release of 1,6-anhydromuropeptides from peptidoglycan. Lytic transglycosylase catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do goose-type lysozymes. However, in addition, it also makes a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCR34480.1 | 5.46e-120 | 3 | 225 | 7 | 229 |
| QDP99068.1 | 7.11e-115 | 1 | 226 | 1 | 227 |
| QPQ35348.1 | 2.26e-114 | 1 | 226 | 1 | 227 |
| AWE07400.1 | 5.74e-112 | 5 | 226 | 4 | 226 |
| ATP40536.1 | 1.59e-111 | 1 | 226 | 1 | 227 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
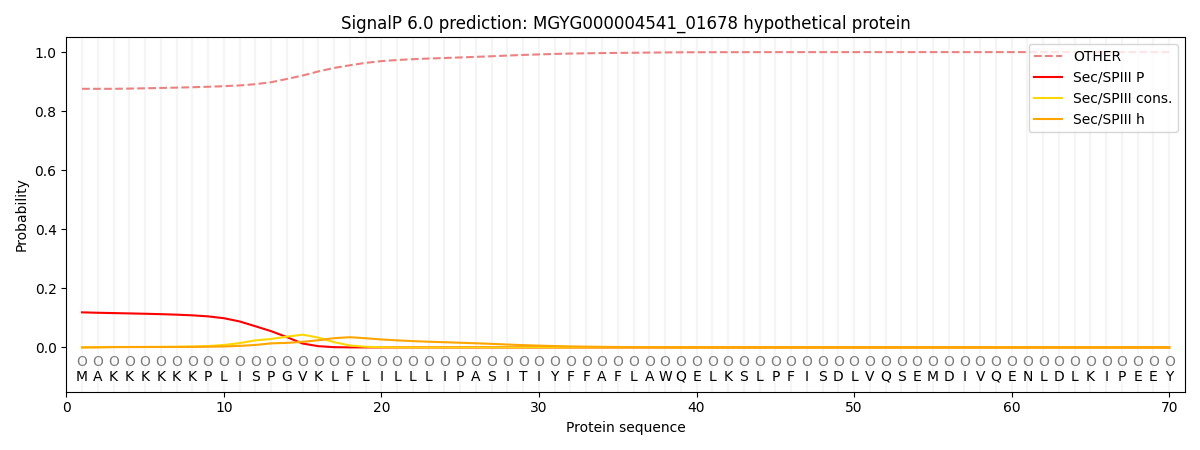
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.875946 | 0.003109 | 0.002050 | 0.000030 | 0.000013 | 0.118877 |