You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004547_01786
You are here: Home > Sequence: MGYG000004547_01786
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; ; | |||||||||||
| CAZyme ID | MGYG000004547_01786 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 221; End: 3760 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 97 | 299 | 2.9e-64 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14256 | Dockerin_I | 2.76e-07 | 1110 | 1170 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam17957 | Big_7 | 2.59e-06 | 519 | 586 | 1 | 67 | Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
| smart00656 | Amb_all | 0.001 | 99 | 303 | 3 | 190 | Amb_all domain. |
| pfam18884 | TSP3_bac | 0.005 | 463 | 484 | 1 | 22 | Bacterial TSP3 repeat. This entry contains a novel bacterial thrombospondin type 3 repeat which differs from the typical consensus by containing a glutamate in place of one of the calcium binding aspartate residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYR21100.1 | 1.10e-173 | 35 | 821 | 41 | 835 |
| AEI43214.1 | 3.40e-171 | 30 | 818 | 31 | 760 |
| AFH63199.1 | 4.78e-171 | 30 | 818 | 31 | 760 |
| AFC30874.1 | 4.78e-171 | 30 | 818 | 31 | 760 |
| AWV33235.1 | 6.42e-168 | 41 | 819 | 54 | 840 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 4.59e-43 | 41 | 513 | 21 | 419 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q5B297 | 8.78e-43 | 35 | 513 | 17 | 416 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 9.44e-42 | 41 | 512 | 21 | 418 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| Q2UB83 | 4.27e-41 | 41 | 513 | 21 | 419 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| A1DPF0 | 1.21e-39 | 41 | 512 | 22 | 419 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
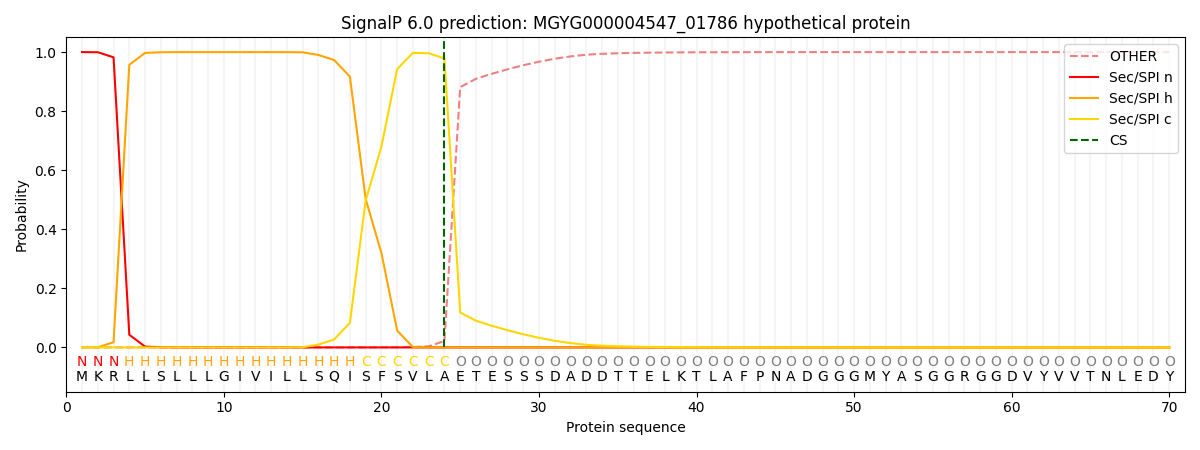
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000361 | 0.998785 | 0.000282 | 0.000204 | 0.000181 | 0.000176 |
