You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004547_01911
You are here: Home > Sequence: MGYG000004547_01911
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; ; | |||||||||||
| CAZyme ID | MGYG000004547_01911 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 283; End: 5061 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 98 | 296 | 9.2e-69 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 7.97e-05 | 2 | 407 | 1 | 341 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 0.003 | 96 | 296 | 1 | 186 | Amb_all domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19050.1 | 1.17e-309 | 1 | 1497 | 7 | 1522 |
| AUO19476.1 | 6.75e-99 | 35 | 532 | 30 | 491 |
| AUO19400.1 | 1.70e-95 | 37 | 524 | 37 | 507 |
| AUO19402.1 | 9.01e-93 | 42 | 517 | 32 | 494 |
| QEL04270.1 | 1.02e-91 | 37 | 520 | 10 | 469 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 7.51e-52 | 40 | 520 | 20 | 413 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 5.30e-50 | 40 | 520 | 20 | 413 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q5B297 | 5.56e-50 | 40 | 520 | 20 | 410 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 2.42e-49 | 40 | 520 | 20 | 413 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| A1DPF0 | 3.36e-49 | 40 | 520 | 21 | 414 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
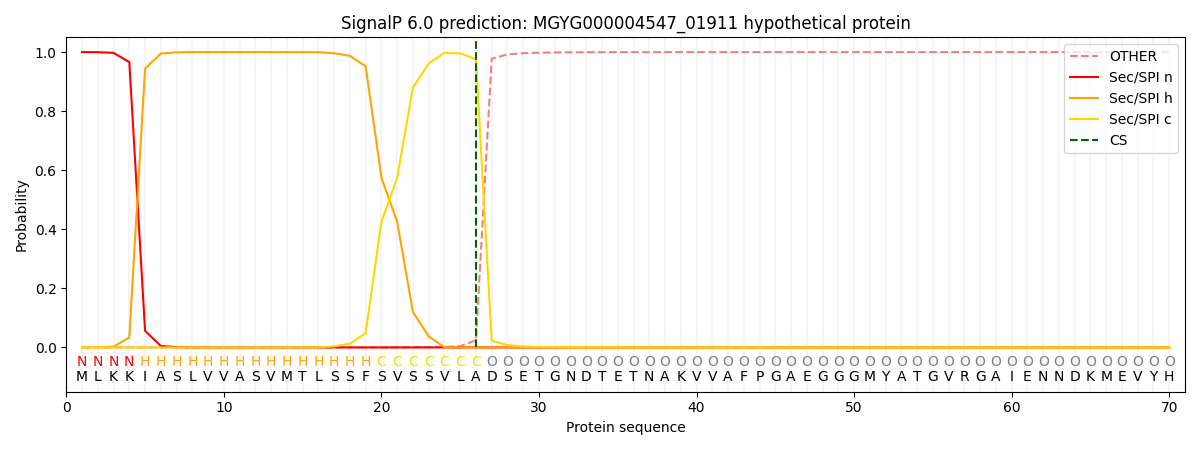
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000276 | 0.998932 | 0.000220 | 0.000205 | 0.000185 | 0.000159 |
