You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004561_00548
You are here: Home > Sequence: MGYG000004561_00548
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002480935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002480935 | |||||||||||
| CAZyme ID | MGYG000004561_00548 | |||||||||||
| CAZy Family | GH143 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7767; End: 11129 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH143 | 23 | 568 | 6.7e-256 | 0.9910071942446043 |
| GH142 | 631 | 1114 | 5.9e-199 | 0.9958246346555324 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13088 | BNR_2 | 2.68e-07 | 267 | 341 | 168 | 247 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG3408 | GDB1 | 4.43e-05 | 746 | 964 | 348 | 551 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| PRK10137 | PRK10137 | 0.003 | 826 | 950 | 581 | 712 | alpha-glucosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85039.1 | 0.0 | 21 | 1116 | 16 | 1108 |
| QUT58490.1 | 0.0 | 23 | 1116 | 19 | 1097 |
| QEW37190.1 | 0.0 | 23 | 1116 | 19 | 1097 |
| QQY37484.1 | 0.0 | 23 | 1116 | 19 | 1097 |
| ABR40380.1 | 0.0 | 23 | 1116 | 19 | 1097 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQR_A | 0.0 | 31 | 1116 | 30 | 1101 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQS_A | 0.0 | 31 | 1116 | 30 | 1101 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 6M5A_A | 4.25e-24 | 651 | 1116 | 304 | 809 | Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 3.90e-23 | 651 | 1116 | 335 | 840 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
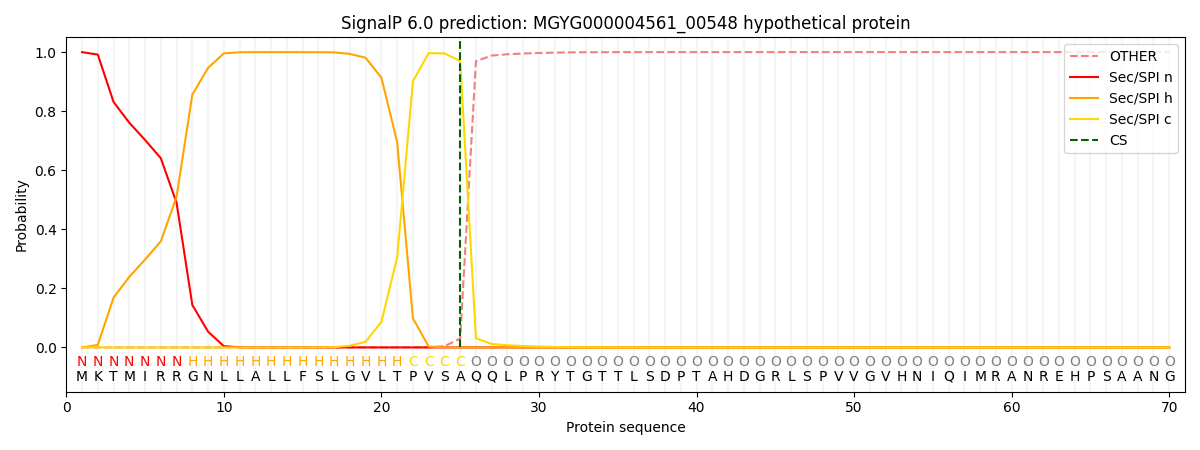
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000477 | 0.998608 | 0.000270 | 0.000249 | 0.000198 | 0.000151 |
