You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004561_00739
You are here: Home > Sequence: MGYG000004561_00739
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002480935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002480935 | |||||||||||
| CAZyme ID | MGYG000004561_00739 | |||||||||||
| CAZy Family | PL10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75250; End: 77148 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL10 | 325 | 616 | 3.4e-112 | 0.9930555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09492 | Pec_lyase | 3.87e-127 | 325 | 618 | 1 | 289 | Pectic acid lyase. Members of this family are isozymes of pectate lyase (EC:4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. |
| TIGR02474 | pec_lyase | 7.67e-112 | 325 | 618 | 1 | 290 | pectate lyase, PelA/Pel-15E family. Members of this family are isozymes of pectate lyase (EC 4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. [Energy metabolism, Biosynthesis and degradation of polysaccharides] |
| COG0657 | Aes | 2.07e-24 | 12 | 278 | 20 | 308 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 5.53e-16 | 109 | 257 | 37 | 207 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| pfam00326 | Peptidase_S9 | 2.27e-13 | 141 | 279 | 65 | 208 | Prolyl oligopeptidase family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ01761.1 | 1.53e-103 | 291 | 629 | 34 | 374 |
| AKP52260.1 | 2.24e-102 | 280 | 629 | 22 | 372 |
| QEC53563.1 | 2.32e-102 | 295 | 624 | 38 | 365 |
| SOE21140.1 | 8.26e-101 | 285 | 627 | 14 | 356 |
| AFO38207.1 | 2.08e-99 | 295 | 628 | 30 | 362 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R76_A | 1.40e-61 | 305 | 627 | 74 | 407 | ChainA, pectate lyase [Niveispirillum irakense] |
| 7DWC_A | 1.45e-58 | 50 | 280 | 32 | 264 | ChainA, Xylanase [Bacteroides thetaiotaomicron VPI-5482],7DWC_B Chain B, Xylanase [Bacteroides thetaiotaomicron VPI-5482],7DWC_C Chain C, Xylanase [Bacteroides thetaiotaomicron VPI-5482],7DWC_D Chain D, Xylanase [Bacteroides thetaiotaomicron VPI-5482] |
| 1GXM_A | 4.03e-51 | 321 | 618 | 40 | 323 | Family10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXM_B Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXN_A Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus] |
| 1GXO_A | 5.53e-50 | 321 | 618 | 40 | 323 | MutantD189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid [Cellvibrio japonicus] |
| 6A6O_A | 6.58e-38 | 73 | 281 | 57 | 280 | ChainA, Esterase/lipase-like protein [Caldicellulosiruptor lactoaceticus 6A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EV35 | 2.63e-44 | 68 | 282 | 45 | 268 | Acetylxylan esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axeA1 PE=1 SV=1 |
| P26223 | 1.57e-17 | 73 | 262 | 405 | 612 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
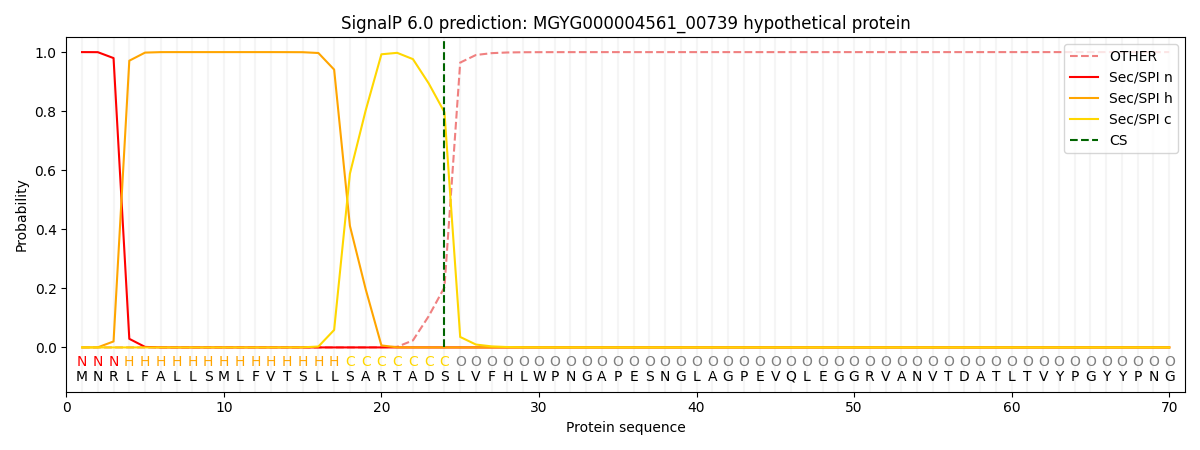
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000187 | 0.999206 | 0.000155 | 0.000154 | 0.000141 | 0.000135 |
