You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004562_01175
You are here: Home > Sequence: MGYG000004562_01175
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gemella morbillorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Gemellaceae; Gemella; Gemella morbillorum | |||||||||||
| CAZyme ID | MGYG000004562_01175 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49632; End: 50552 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK06347 | PRK06347 | 3.67e-15 | 26 | 129 | 478 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 3.74e-15 | 26 | 160 | 404 | 554 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 1.23e-14 | 25 | 161 | 328 | 481 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK13914 | PRK13914 | 2.18e-14 | 26 | 286 | 198 | 460 | invasion associated endopeptidase. |
| pfam00877 | NLPC_P60 | 1.06e-13 | 217 | 306 | 15 | 104 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SQH56165.1 | 2.02e-169 | 1 | 306 | 1 | 328 |
| QGS08598.1 | 5.11e-167 | 1 | 306 | 1 | 326 |
| AME08782.1 | 8.88e-145 | 1 | 306 | 1 | 308 |
| AXI26353.1 | 1.58e-142 | 1 | 306 | 1 | 336 |
| VEI39452.1 | 1.42e-134 | 1 | 306 | 1 | 370 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4B8V_A | 7.81e-07 | 30 | 142 | 44 | 173 | ChainA, Extracellular Protein 6 [Fulvia fulva],4B9H_A Chain A, Extracellular Protein 6 [Fulvia fulva] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34669 | 3.36e-16 | 12 | 135 | 12 | 127 | Cell wall-binding protein YocH OS=Bacillus subtilis (strain 168) OX=224308 GN=yocH PE=1 SV=1 |
| P54421 | 3.06e-14 | 7 | 134 | 6 | 134 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| Q49UX4 | 5.34e-14 | 6 | 135 | 4 | 135 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
| Q01835 | 5.86e-14 | 5 | 286 | 154 | 490 | Probable endopeptidase p60 OS=Listeria grayi OX=1641 GN=iap PE=3 SV=1 |
| Q6GJK9 | 7.65e-14 | 18 | 139 | 17 | 144 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
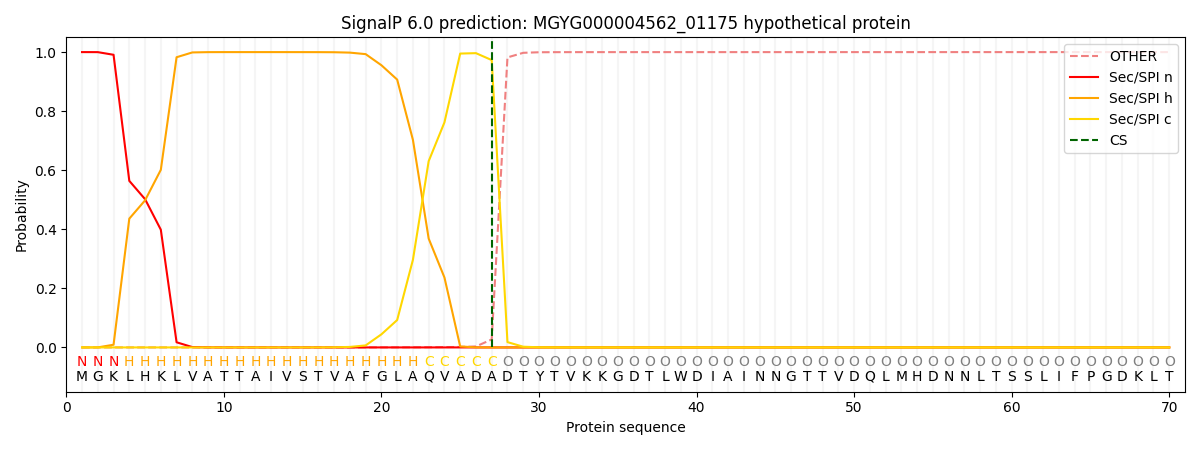
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000226 | 0.999056 | 0.000175 | 0.000177 | 0.000174 | 0.000153 |
