You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004572_01452
You are here: Home > Sequence: MGYG000004572_01452
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
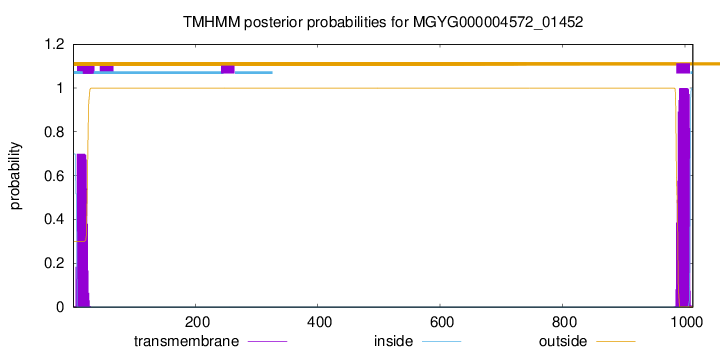
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; | |||||||||||
| CAZyme ID | MGYG000004572_01452 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 430; End: 3471 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 8.49e-28 | 224 | 402 | 50 | 236 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 1.11e-13 | 278 | 344 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| smart00047 | LYZ2 | 6.28e-12 | 263 | 396 | 2 | 141 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| COG1705 | FlgJ | 1.70e-06 | 283 | 405 | 56 | 195 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| pfam05297 | Herpes_LMP1 | 4.07e-05 | 850 | 948 | 269 | 379 | Herpesvirus latent membrane protein 1 (LMP1). This family consists of several latent membrane protein 1 or LMP1s mostly from Epstein-Barr virus. LMP1 of EBV is a 62-65 kDa plasma membrane protein possessing six membrane spanning regions, a short cytoplasmic N-terminus and a long cytoplasmic carboxy tail of 200 amino acids. EBV latent membrane protein 1 (LMP1) is essential for EBV-mediated transformation and has been associated with several cases of malignancies. EBV-like viruses in Cynomolgus monkeys (Macaca fascicularis) have been associated with high lymphoma rates in immunosuppressed monkeys |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK17141.1 | 6.66e-258 | 2 | 862 | 3 | 859 |
| AWY97555.1 | 1.59e-188 | 1 | 709 | 1 | 717 |
| QOV20091.1 | 5.41e-129 | 48 | 497 | 40 | 477 |
| QQV05342.1 | 9.00e-107 | 23 | 499 | 69 | 539 |
| QPS13527.1 | 1.15e-101 | 53 | 557 | 76 | 568 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 4.88e-31 | 108 | 402 | 31 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 5.01e-29 | 108 | 402 | 399 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 6.26e-29 | 108 | 402 | 443 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| P39848 | 9.60e-06 | 232 | 394 | 694 | 863 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
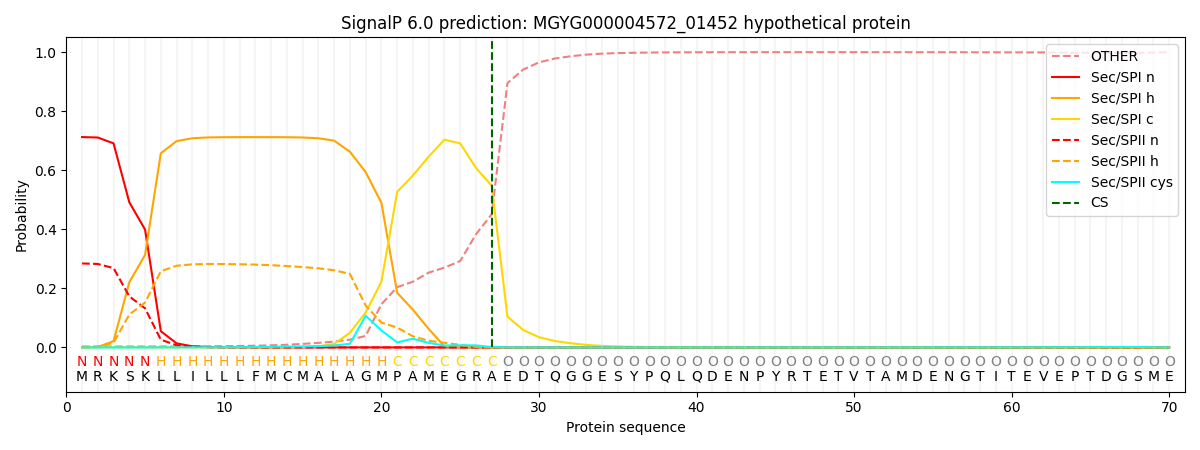
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006041 | 0.700547 | 0.291786 | 0.000909 | 0.000407 | 0.000294 |