You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004587_00434
You are here: Home > Sequence: MGYG000004587_00434
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
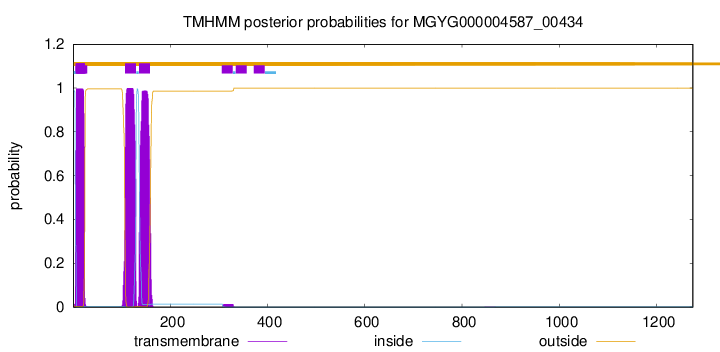
TMHMM annotations
Basic Information help
| Species | UBA1409 sp900553675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1409; UBA1409 sp900553675 | |||||||||||
| CAZyme ID | MGYG000004587_00434 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97056; End: 100883 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 196 | 1013 | 5.7e-162 | 0.9440459110473458 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 3.66e-125 | 354 | 795 | 3 | 333 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| pfam17829 | GH115_C | 2.18e-40 | 1080 | 1254 | 1 | 172 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
| cd04079 | CBM6_agarase-like | 2.39e-05 | 1134 | 1224 | 42 | 115 | Carbohydrate Binding Module 6 (CBM6); appended mainly to glycoside hydrolase (GH) family 16 alpha- and beta agarases. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family 16 agarases. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the activity of alpha- and beta-agarase catalytic modules which are involved in the hydrolysis of 1,4-beta-D-galactosidic linkages. These CBM6s bind specifically to the non-reducing end of agarose chains, recognizing only the first repeat of the disaccharide, and directing the appended catalytic modules to areas of the plant cell wall attacked by beta-agarases. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. This family includes three tandem CBM6s from the Saccharophagus degradans agarase Aga86E, and three tandem CBM6s from Vibrio sp. strain PO-303 AgaA; in both these proteins these are appended to a GH16 domain. Vibrio AgaA also contains a Big-2-like protein-protein interaction domain. This family also includes two tandem CBM6s from an endo-type beta-agarase from a deep-sea Microbulbifer-like isolate, which are appended to a GH16 domain, and two of three CBM6s of Alteromonas agarilytica AgaA alpha-agarase, which are appended to a GH96 domain. |
| pfam03648 | Glyco_hydro_67N | 1.21e-04 | 231 | 315 | 44 | 119 | Glycosyl hydrolase family 67 N-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the N-terminal region of alpha-glucuronidase. The N-terminal domain forms a two-layer sandwich, each layer being formed by a beta sheet of five strands. A further two helices form part of the interface with the central, catalytic, module (pfam07488). |
| cd14489 | CBM_SBP_bac_1_like | 0.002 | 1134 | 1224 | 52 | 134 | Putative Carbohydrate Binding Module (CBM) of extracellular solute-binding protein family 1. Domains in this family co-occur with extracellular solute-binding domains which are periplasmic components of ABC-type sugar transport systems involved in carbohydrate transport and metabolism. Carbohydrate binding modules of family 6 (CBM6), also known as cellulose binding domain family VI (CBD VI), and related CBMs (CBM35 and CBM36) are non-catalytic carbohydrate binding domains found in a range of enzymes that display activities against a diverse range of carbohydrate targets, including mannan, xylan, beta-glucans, cellulose, agarose, and arabinans. These domains facilitate the strong binding of co-occuring (catalytic) modules to their insoluble substrates. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNU66651.1 | 1.03e-220 | 194 | 1256 | 35 | 982 |
| AQR97306.1 | 2.30e-205 | 197 | 1253 | 4 | 940 |
| AGF58615.1 | 3.23e-205 | 197 | 1253 | 4 | 940 |
| AWV33044.1 | 7.77e-204 | 199 | 1259 | 15 | 955 |
| AIQ23284.1 | 9.27e-202 | 196 | 1258 | 5 | 947 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NPS_A | 9.73e-156 | 216 | 1256 | 28 | 964 | Crystalstructure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112],6NPS_B Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112] |
| 4ZMH_A | 4.95e-111 | 197 | 1255 | 14 | 933 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
| 7PUG_A | 1.59e-84 | 193 | 957 | 13 | 646 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 4C90_A | 2.06e-84 | 207 | 957 | 54 | 661 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 7PXQ_A | 3.45e-83 | 193 | 957 | 12 | 645 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
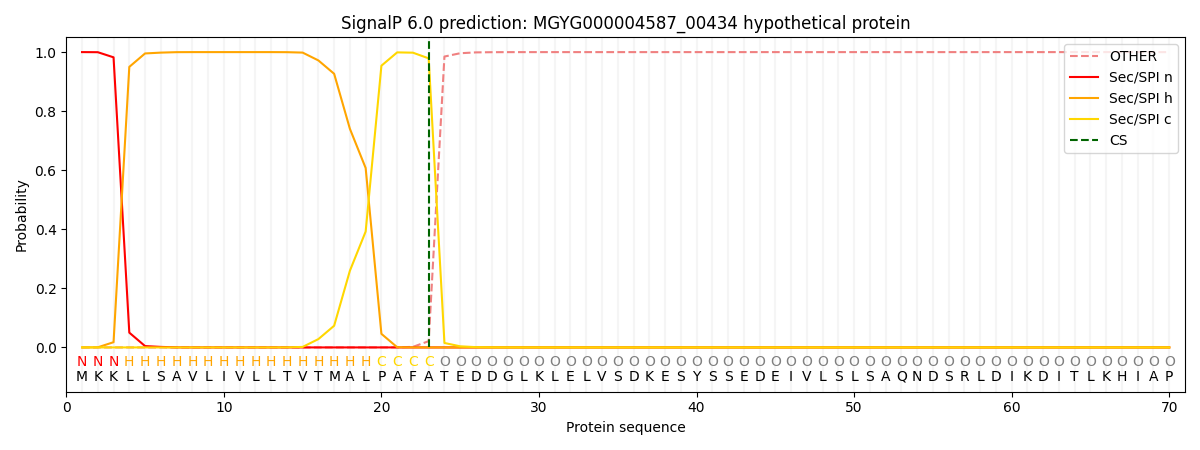
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000249 | 0.998966 | 0.000206 | 0.000186 | 0.000179 | 0.000167 |