You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004588_01026
You are here: Home > Sequence: MGYG000004588_01026
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900317685 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900317685 | |||||||||||
| CAZyme ID | MGYG000004588_01026 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35341; End: 37875 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 249 | 511 | 9.8e-47 | 0.9537037037037037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 4.17e-27 | 208 | 549 | 21 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 2.57e-25 | 275 | 794 | 115 | 600 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 2.28e-10 | 269 | 791 | 103 | 586 | Probable beta-xylosidase; Provisional |
| pfam01915 | Glyco_hydro_3_C | 5.07e-09 | 589 | 830 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIX66430.1 | 0.0 | 5 | 843 | 17 | 776 |
| QIA06568.1 | 0.0 | 6 | 843 | 14 | 779 |
| QDK80972.1 | 0.0 | 1 | 844 | 1 | 773 |
| QEC66867.1 | 0.0 | 1 | 843 | 1 | 772 |
| QJW89827.1 | 0.0 | 6 | 844 | 5 | 773 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JP0_A | 1.14e-27 | 250 | 830 | 101 | 629 | Bacteroidesovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus],5JP0_B Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus] |
| 5Z87_A | 2.71e-27 | 240 | 793 | 112 | 629 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 5Z9S_A | 9.50e-25 | 201 | 794 | 49 | 625 | Functionaland Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum],5Z9S_B Functional and Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum] |
| 5M6G_A | 4.02e-24 | 278 | 830 | 141 | 614 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 4ZOA_A | 2.34e-23 | 240 | 794 | 73 | 555 | CrystalStructure of beta-glucosidase from Listeria innocua in complex with isofagomine [Listeria innocua Clip11262],4ZOA_B Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine [Listeria innocua Clip11262],4ZOB_A Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone [Listeria innocua Clip11262],4ZOB_B Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone [Listeria innocua Clip11262],4ZOD_A Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose [Listeria innocua Clip11262],4ZOD_B Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose [Listeria innocua Clip11262],4ZOE_A Crystal Structure of beta-glucosidase from Listeria innocua [Listeria innocua Clip11262],4ZOE_B Crystal Structure of beta-glucosidase from Listeria innocua [Listeria innocua Clip11262] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 1.49e-94 | 51 | 810 | 35 | 640 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| B8NGU6 | 3.97e-87 | 199 | 825 | 88 | 623 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q2UFP8 | 4.45e-85 | 199 | 825 | 92 | 627 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q5BCC6 | 2.01e-80 | 223 | 810 | 98 | 601 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| A7LXU3 | 6.44e-27 | 250 | 830 | 123 | 651 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
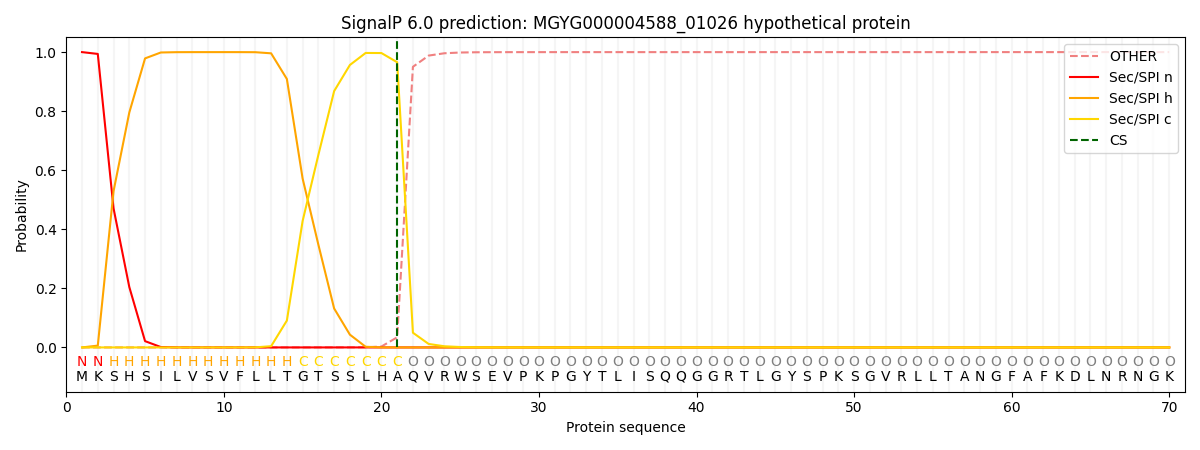
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000318 | 0.998986 | 0.000192 | 0.000172 | 0.000150 | 0.000143 |
