You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004599_00123
You are here: Home > Sequence: MGYG000004599_00123
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; | |||||||||||
| CAZyme ID | MGYG000004599_00123 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | Isomalto-dextranase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52394; End: 53854 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 207 | 461 | 3.8e-31 | 0.9388646288209607 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17801 | Melibiase_C | 1.18e-14 | 412 | 482 | 5 | 74 | Alpha galactosidase C-terminal beta sandwich domain. This domain is found at the C-terminus of alpha galactosidase enzymes. |
| PLN02229 | PLN02229 | 1.47e-08 | 258 | 483 | 205 | 418 | alpha-galactosidase |
| PLN02808 | PLN02808 | 7.21e-08 | 349 | 483 | 249 | 384 | alpha-galactosidase |
| PLN02692 | PLN02692 | 2.28e-04 | 358 | 479 | 282 | 405 | alpha-galactosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT44530.1 | 6.79e-185 | 9 | 484 | 9 | 492 |
| QUT79686.1 | 6.79e-185 | 9 | 484 | 9 | 492 |
| QUU07210.1 | 6.79e-185 | 9 | 484 | 9 | 492 |
| QRM98947.1 | 2.74e-184 | 9 | 484 | 9 | 492 |
| QNT67171.1 | 1.21e-168 | 11 | 482 | 11 | 473 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AWO_A | 2.21e-81 | 58 | 483 | 38 | 466 | Arthrobacterglobiformis T6 isomalto-dextranse [Arthrobacter globiformis],5AWP_A Arthrobacter globiformis T6 isomalto-dextranase complexed with isomaltose [Arthrobacter globiformis],5AWQ_A Arthrobacter globiformis T6 isomalto-dextranse complexed with panose [Arthrobacter globiformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q44052 | 8.33e-82 | 58 | 483 | 64 | 492 | Isomalto-dextranase OS=Arthrobacter globiformis OX=1665 GN=imd PE=1 SV=1 |
| Q5AX28 | 1.72e-06 | 313 | 485 | 228 | 402 | Alpha-galactosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aglD PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
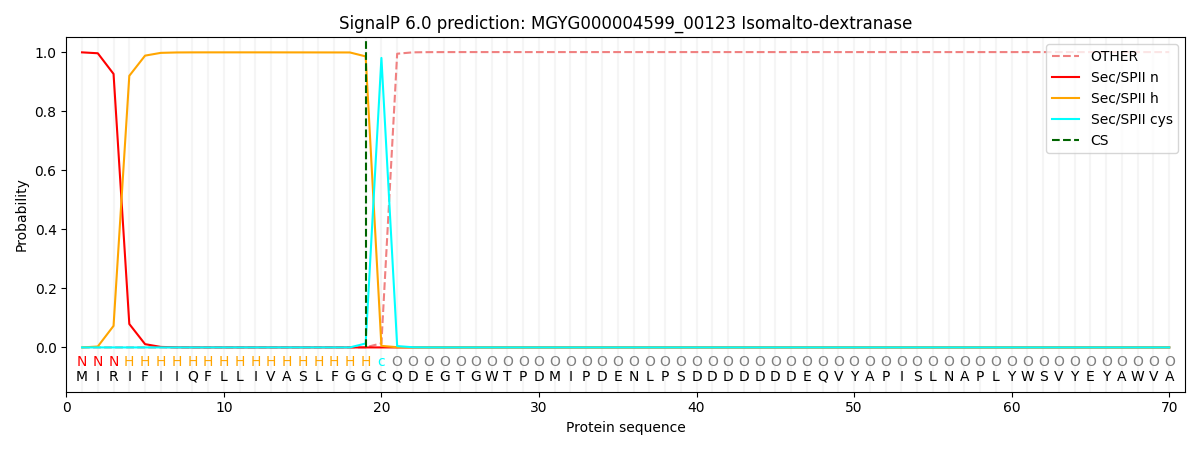
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000874 | 0.999181 | 0.000000 | 0.000000 | 0.000000 |
