You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004599_01453
You are here: Home > Sequence: MGYG000004599_01453
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; | |||||||||||
| CAZyme ID | MGYG000004599_01453 | |||||||||||
| CAZy Family | GH66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7549; End: 9336 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH66 | 40 | 593 | 6.1e-168 | 0.9928057553956835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13199 | Glyco_hydro_66 | 0.0 | 42 | 591 | 1 | 557 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
| cd14745 | GH66 | 1.21e-152 | 127 | 458 | 2 | 331 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU93474.1 | 1.60e-295 | 1 | 595 | 1 | 588 |
| AHF13484.1 | 8.34e-258 | 1 | 594 | 1 | 590 |
| QIK54447.1 | 5.40e-201 | 12 | 595 | 14 | 587 |
| QIK59838.1 | 8.13e-198 | 12 | 595 | 14 | 587 |
| ASF42017.1 | 1.76e-197 | 1 | 594 | 1 | 579 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AXG_A | 3.66e-113 | 23 | 593 | 32 | 609 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXG_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223] |
| 5AXH_A | 4.01e-112 | 23 | 593 | 32 | 609 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXH_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose [Thermoanaerobacter pseudethanolicus ATCC 33223] |
| 5X7G_A | 1.02e-61 | 42 | 590 | 31 | 718 | CrystalStructure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase [Paenibacillus sp. 598K],5X7H_A Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose [Paenibacillus sp. 598K] |
| 3WNK_A | 1.89e-55 | 42 | 590 | 32 | 719 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNL_A | 2.09e-54 | 42 | 590 | 13 | 700 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94286 | 1.11e-55 | 2 | 593 | 11 | 739 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
| P70873 | 2.63e-53 | 34 | 593 | 34 | 731 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 GN=cit PE=3 SV=1 |
| Q54443 | 3.03e-21 | 38 | 592 | 106 | 732 | Dextranase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=dexA PE=1 SV=2 |
| P39653 | 2.80e-20 | 21 | 592 | 147 | 800 | Dextranase OS=Streptococcus downei OX=1317 GN=dex PE=1 SV=1 |
| Q59979 | 3.26e-18 | 83 | 593 | 37 | 598 | Dextranase OS=Streptococcus salivarius OX=1304 GN=dex PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
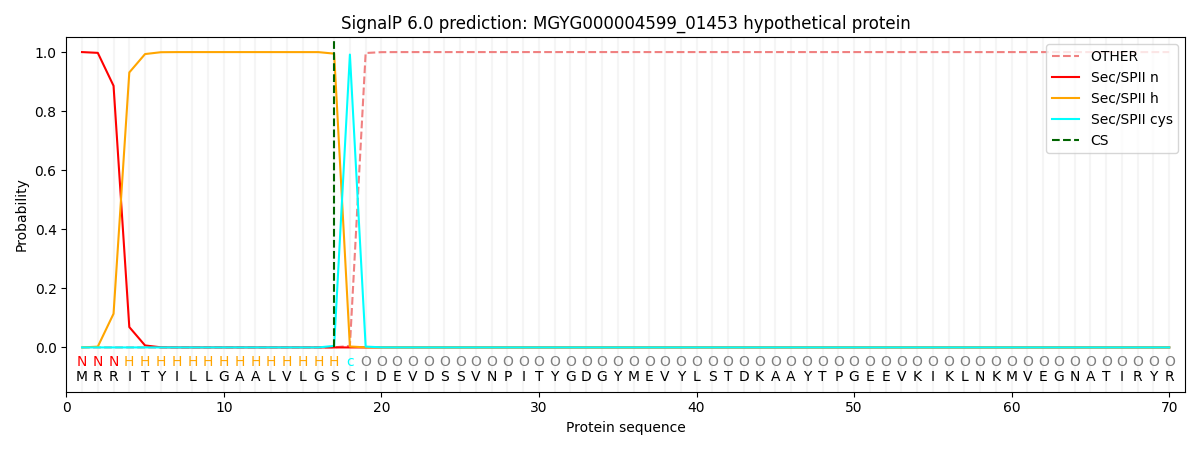
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000003 | 1.000087 | 0.000000 | 0.000000 | 0.000000 |
