You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004605_02234
You are here: Home > Sequence: MGYG000004605_02234
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
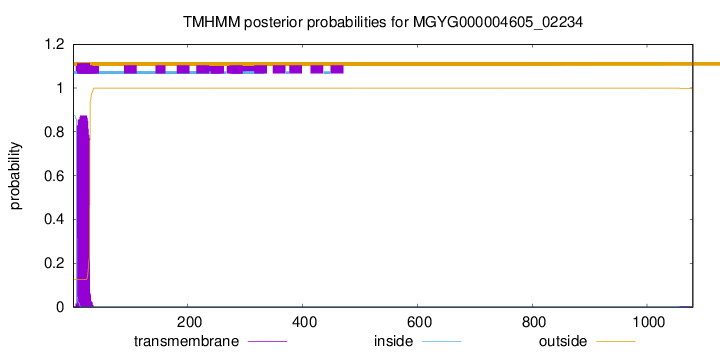
TMHMM annotations
Basic Information help
| Species | UBA7173 sp900555165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; UBA7173 sp900555165 | |||||||||||
| CAZyme ID | MGYG000004605_02234 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3282; End: 6524 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 861 | 1005 | 2e-24 | 0.7287234042553191 |
| CBM13 | 386 | 533 | 1.1e-23 | 0.7340425531914894 |
| CBM13 | 623 | 766 | 7.8e-20 | 0.7127659574468085 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 6.02e-18 | 422 | 511 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 8.92e-17 | 385 | 464 | 12 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.42e-16 | 898 | 986 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 2.14e-16 | 857 | 940 | 8 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.57e-13 | 660 | 759 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHS59416.1 | 4.90e-122 | 1 | 456 | 1 | 454 |
| ACU61780.1 | 1.24e-119 | 1 | 450 | 1 | 450 |
| QNN78011.1 | 1.15e-51 | 23 | 287 | 28 | 296 |
| ABX42085.1 | 1.38e-51 | 23 | 286 | 37 | 300 |
| AWV08387.1 | 1.88e-50 | 23 | 270 | 15 | 261 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3HZB_A | 6.68e-18 | 773 | 858 | 4 | 90 | Crystalstructure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_B Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_C Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_D Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_E Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_F Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_G Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101],3HZB_H Crystal structure of a betagamma-crystallin domain from Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101] |
| 3SNZ_A | 1.35e-16 | 769 | 859 | 3 | 94 | Crystalstructure of a mutant W39D of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3I9H_A | 5.99e-15 | 769 | 852 | 1 | 85 | Crystalstructure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_B Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_C Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_D Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_E Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_F Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_G Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_H Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SNY_A | 7.72e-15 | 769 | 859 | 3 | 94 | Crystalstructure of a mutant T82R of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SO0_A | 1.05e-14 | 769 | 859 | 3 | 94 | Crystalstructure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_B Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_C Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_D Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_E Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_F Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_G Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_H Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P02966 | 1.03e-12 | 503 | 614 | 67 | 172 | Development-specific protein S OS=Myxococcus xanthus OX=34 GN=tps PE=1 SV=1 |
| P02967 | 5.00e-12 | 503 | 614 | 67 | 173 | Development-specific protein S homolog OS=Myxococcus xanthus OX=34 GN=ops PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
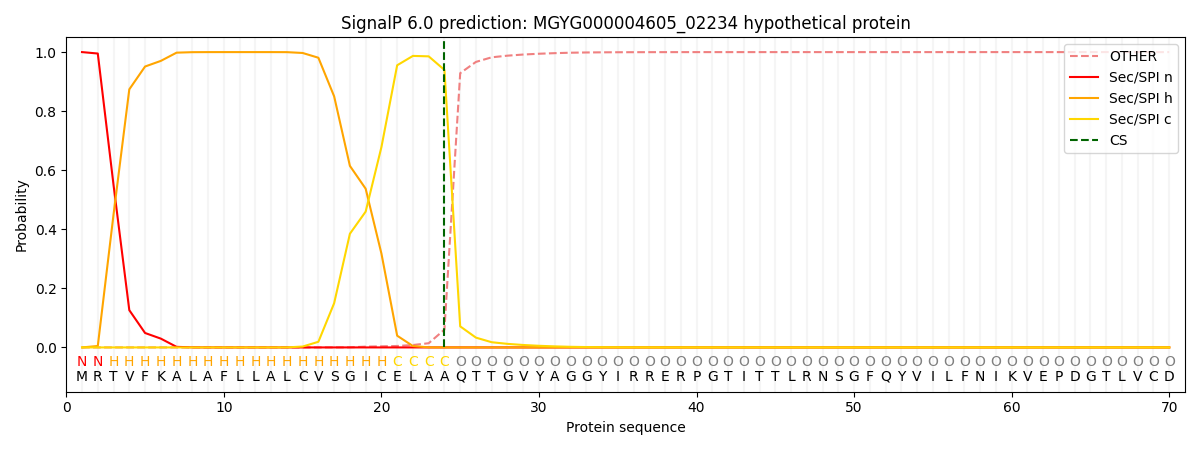
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000317 | 0.999047 | 0.000165 | 0.000168 | 0.000144 | 0.000139 |