You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004609_01304
You are here: Home > Sequence: MGYG000004609_01304
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
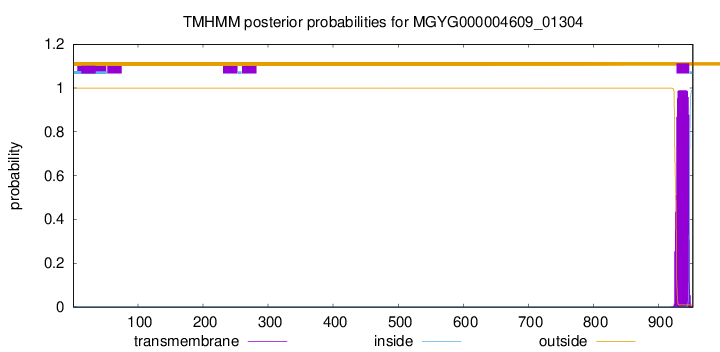
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; | |||||||||||
| CAZyme ID | MGYG000004609_01304 | |||||||||||
| CAZy Family | GH146 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 79; End: 2940 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 1 | 341 | 1.3e-113 | 0.6381709741550696 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 1.03e-89 | 1 | 341 | 180 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 1.38e-64 | 1 | 345 | 183 | 505 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| pfam07523 | Big_3 | 3.06e-14 | 792 | 858 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07523 | Big_3 | 1.62e-11 | 709 | 775 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam12733 | Cadherin-like | 3.20e-08 | 609 | 688 | 5 | 86 | Cadherin-like beta sandwich domain. This domain is found in several bacterial, metazoan and chlorophyte algal proteins. A profile-profile comparison recovered the cadherin domain and a comparison of the predicted structure of this domain with the crystal structure of the cadherin showed a congruent seven stranded secondary structure. The domain is widespread in bacteria and seen in the firmicutes, actinobacteria, certain proteobacteria, bacteroides and chlamydiae with an expansion in Clostridium. In contrast, it is limited in its distribution in eukaryotes suggesting that it was derived through lateral transfer from bacteria. In prokaryotes, this domain is widely fused to other domains such as FNIII (Fibronectin Type III), TIG, SLH (S-layer homology), discoidin, cell-wall-binding repeat domain and alpha-amylase-like glycohydrolases. These associations are suggestive of a carbohydrate-binding function for this cadherin-like domain. In animal proteins it is associated with an ATP-grasp domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDP74899.1 | 2.27e-223 | 1 | 689 | 571 | 1291 |
| ARU52215.1 | 3.19e-223 | 1 | 689 | 571 | 1291 |
| QUB98427.1 | 3.19e-223 | 1 | 689 | 571 | 1291 |
| ARK06383.1 | 4.49e-223 | 1 | 689 | 571 | 1291 |
| QJW37681.1 | 2.47e-222 | 1 | 689 | 571 | 1291 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YQH_AAA | 4.81e-64 | 1 | 545 | 219 | 759 | ChainAAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
| 5OPJ_A | 1.19e-61 | 1 | 545 | 219 | 759 | Beta-L-arabinofuranosidase[Bacteroides thetaiotaomicron] |
| 6EX6_A | 5.84e-14 | 83 | 343 | 270 | 547 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 4FD0_A | 7.24e-06 | 710 | 776 | 35 | 102 | Crystalstructure of a putative cell surface protein (BACCAC_03700) from Bacteroides caccae ATCC 43185 at 2.07 A resolution [Bacteroides caccae ATCC 43185] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 2.51e-14 | 668 | 859 | 1265 | 1452 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
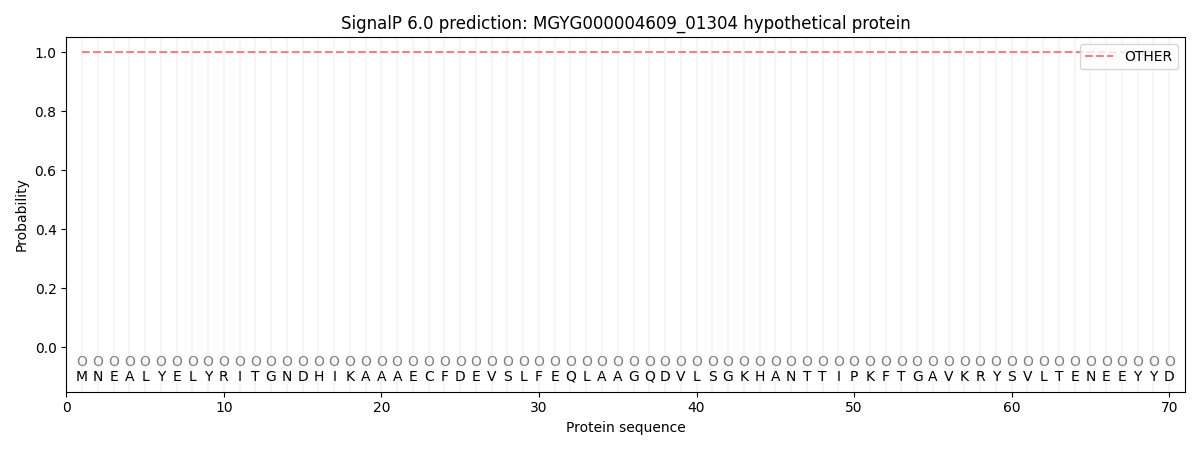
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000049 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |