You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004621_00032
You are here: Home > Sequence: MGYG000004621_00032
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004621_00032 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43592; End: 44974 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 20 | 214 | 1e-32 | 0.9896907216494846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0657 | Aes | 2.94e-23 | 185 | 434 | 9 | 286 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 4.66e-21 | 263 | 434 | 1 | 205 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| cd01833 | XynB_like | 2.47e-17 | 19 | 214 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| cd00229 | SGNH_hydrolase | 5.21e-12 | 21 | 213 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| pfam13472 | Lipase_GDSL_2 | 7.99e-12 | 24 | 205 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAX78799.1 | 1.67e-110 | 3 | 458 | 13 | 488 |
| ATC65549.1 | 2.84e-100 | 6 | 459 | 25 | 487 |
| QOV88633.1 | 6.92e-52 | 1 | 221 | 11 | 229 |
| QNN20991.1 | 2.13e-49 | 8 | 220 | 18 | 228 |
| QDT00894.1 | 3.12e-27 | 222 | 458 | 27 | 277 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AO9_A | 4.33e-28 | 231 | 458 | 19 | 272 | Thestructure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-native [Thermogutta terrifontis],5AOA_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound [Thermogutta terrifontis],5AOB_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound [Thermogutta terrifontis],5AOC_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound [Thermogutta terrifontis] |
| 7BFN_A | 4.42e-28 | 231 | 458 | 20 | 273 | ChainA, Esterase [Thermogutta terrifontis] |
| 7BFO_A | 2.18e-27 | 231 | 458 | 20 | 273 | ChainA, Esterase [Thermogutta terrifontis],7BFR_A Chain A, Esterase [Thermogutta terrifontis],7BFT_A Chain A, Esterase [Thermogutta terrifontis],7BFU_A Chain A, Esterase [Thermogutta terrifontis],7BFV_A Chain A, Esterase [Thermogutta terrifontis] |
| 2YH2_A | 4.29e-12 | 211 | 415 | 33 | 234 | Pyrobaculumcalidifontis esterase monoclinic form [Pyrobaculum calidifontis],2YH2_B Pyrobaculum calidifontis esterase monoclinic form [Pyrobaculum calidifontis],2YH2_C Pyrobaculum calidifontis esterase monoclinic form [Pyrobaculum calidifontis],2YH2_D Pyrobaculum calidifontis esterase monoclinic form [Pyrobaculum calidifontis],3ZWQ_A Hyperthermophilic Esterase From The Archeon Pyrobaculum Calidifontis [Pyrobaculum calidifontis JCM 11548],3ZWQ_B Hyperthermophilic Esterase From The Archeon Pyrobaculum Calidifontis [Pyrobaculum calidifontis JCM 11548] |
| 6K34_A | 4.70e-11 | 236 | 369 | 69 | 195 | CrystalStructure of DphMB1 [Mycobacterium sp. YC-RL4],6K34_B Crystal Structure of DphMB1 [Mycobacterium sp. YC-RL4],6K34_C Crystal Structure of DphMB1 [Mycobacterium sp. YC-RL4],6K34_D Crystal Structure of DphMB1 [Mycobacterium sp. YC-RL4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P71668 | 8.49e-08 | 240 | 369 | 67 | 194 | Esterase LipI OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipI PE=1 SV=1 |
| A0A0G3FWY4 | 1.82e-07 | 261 | 434 | 74 | 255 | Probable N-octanoylanthranilate hydrolase AqdA1 OS=Rhodococcus erythropolis OX=1833 GN=aqdA1 PE=1 SV=1 |
| Q9US38 | 7.00e-07 | 261 | 445 | 101 | 324 | AB hydrolase superfamily protein C1039.03 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC1039.03 PE=3 SV=1 |
| P24484 | 2.81e-06 | 235 | 352 | 133 | 249 | Lipase 2 OS=Moraxella sp. (strain TA144) OX=77152 GN=lip2 PE=1 SV=1 |
| Q9LMA7 | 4.70e-06 | 241 | 369 | 53 | 196 | Probable carboxylesterase 1 OS=Arabidopsis thaliana OX=3702 GN=CXE1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
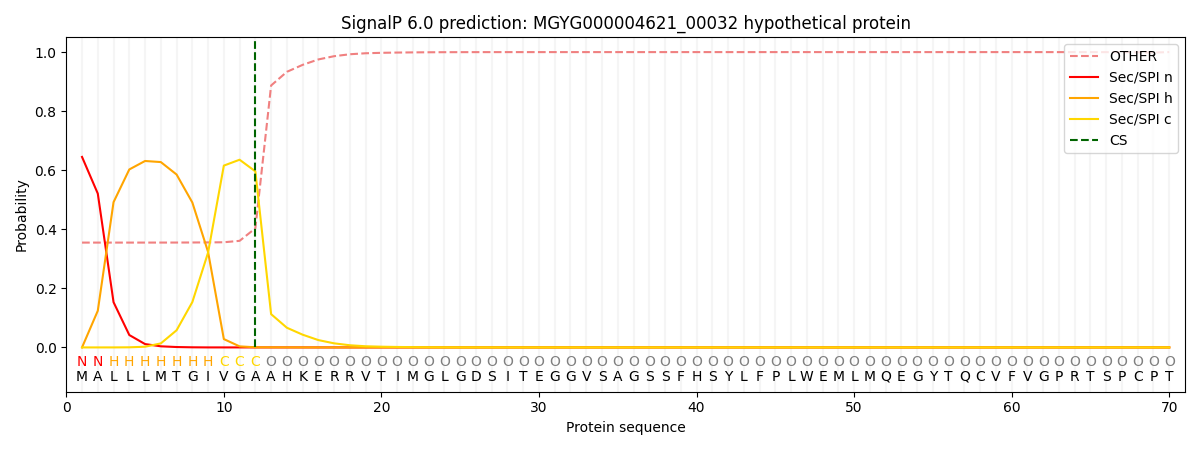
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.361712 | 0.637170 | 0.000250 | 0.000356 | 0.000246 | 0.000254 |
