You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004621_00966
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella;
|
| CAZyme ID |
MGYG000004621_00966
|
| CAZy Family |
GH28 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004621 |
2838817 |
MAG |
France |
Europe |
|
| Gene Location |
Start: 14462;
End: 16051
Strand: +
|
No EC number prediction in MGYG000004621_00966.
| Family |
Start |
End |
Evalue |
family coverage |
| GH28 |
230 |
478 |
4.5e-22 |
0.6523076923076923 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG5434
|
Pgu1 |
2.57e-08 |
256 |
369 |
239 |
345 |
Polygalacturonase [Carbohydrate transport and metabolism]. |
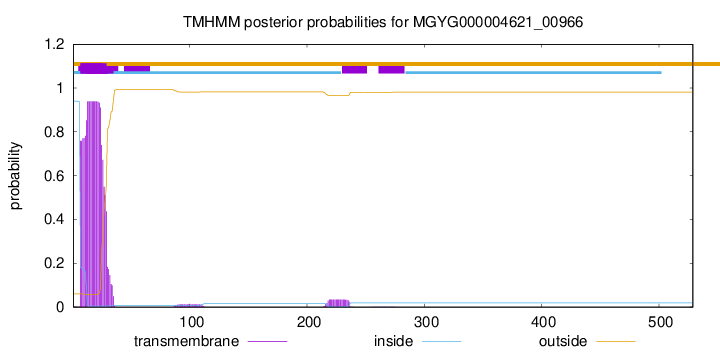
This protein is predicted as SP
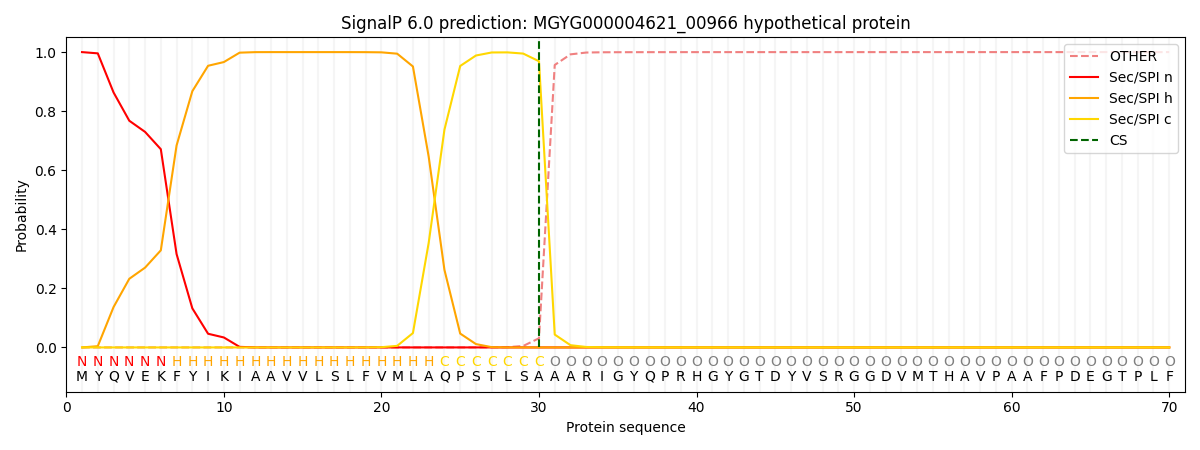
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000336
|
0.998986
|
0.000209
|
0.000155
|
0.000151
|
0.000144
|