You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004621_01102
You are here: Home > Sequence: MGYG000004621_01102
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004621_01102 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11248; End: 12726 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 74 | 446 | 1.6e-41 | 0.8156424581005587 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 2.83e-12 | 183 | 386 | 86 | 255 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 9.58e-08 | 183 | 393 | 92 | 251 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCD21417.1 | 4.90e-174 | 6 | 486 | 1 | 479 |
| QUT64972.1 | 5.42e-168 | 24 | 488 | 17 | 479 |
| BBK85776.1 | 6.00e-168 | 24 | 488 | 20 | 482 |
| QPH58159.1 | 6.00e-168 | 24 | 488 | 20 | 482 |
| QDH77939.1 | 4.39e-167 | 7 | 486 | 5 | 478 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MU9_A | 8.14e-17 | 183 | 459 | 117 | 339 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 6SHD_A | 7.86e-11 | 181 | 375 | 131 | 282 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.36e-09 | 181 | 375 | 132 | 283 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 4C1S_A | 1.71e-09 | 183 | 448 | 107 | 334 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6SHM_A | 3.20e-09 | 187 | 375 | 139 | 283 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
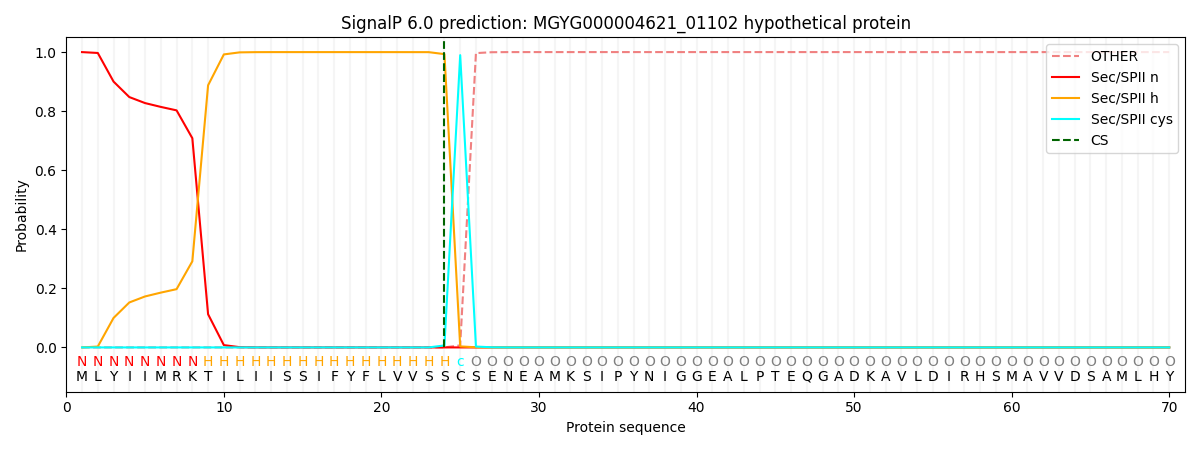
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000012 | 1.000002 | 0.000000 | 0.000000 | 0.000000 |
