You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004627_00850
Basic Information
help
| Species |
Prevotella nigrescens
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella nigrescens
|
| CAZyme ID |
MGYG000004627_00850
|
| CAZy Family |
CE6 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 332 |
|
36826.17 |
9.4998 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004627 |
2663266 |
MAG |
Germany |
Europe |
|
| Gene Location |
Start: 24636;
End: 25634
Strand: -
|
No EC number prediction in MGYG000004627_00850.
MGYG000004627_00850 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
D5EXZ4
|
1.54e-20 |
16 |
238 |
38 |
298 |
Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
This protein is predicted as SP
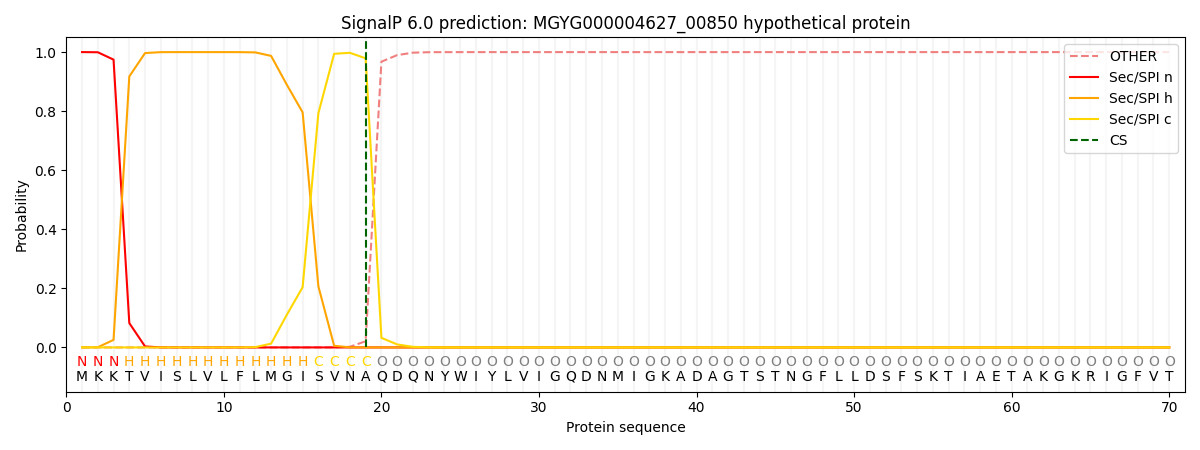
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000352
|
0.998893
|
0.000189
|
0.000187
|
0.000185
|
0.000167
|
There is no transmembrane helices in MGYG000004627_00850.

