You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004632_00474
You are here: Home > Sequence: MGYG000004632_00474
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002300055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002300055 | |||||||||||
| CAZyme ID | MGYG000004632_00474 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 71810; End: 74506 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 1.25e-30 | 698 | 849 | 3 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 6.64e-12 | 445 | 528 | 4 | 81 | Ricin-type beta-trefoil lectin domain-like. |
| cd00110 | LamG | 4.08e-11 | 699 | 838 | 7 | 150 | Laminin G domain; Laminin G-like domains are usually Ca++ mediated receptors that can have binding sites for steroids, beta1 integrins, heparin, sulfatides, fibulin-1, and alpha-dystroglycans. Proteins that contain LamG domains serve a variety of purposes including signal transduction via cell-surface steroid receptors, adhesion, migration and differentiation through mediation of cell adhesion molecules. |
| cd00152 | PTX | 1.57e-09 | 698 | 851 | 16 | 173 | Pentraxins are plasma proteins characterized by their pentameric discoid assembly and their Ca2+ dependent ligand binding, such as Serum amyloid P component (SAP) and C-reactive Protein (CRP), which are cytokine-inducible acute-phase proteins implicated in innate immunity. CRP binds to ligands containing phosphocholine, SAP binds to amyloid fibrils, DNA, chromatin, fibronectin, C4-binding proteins and glycosaminoglycans. "Long" pentraxins have N-terminal extensions to the common pentraxin domain; one group, the neuronal pentraxins, may be involved in synapse formation and remodeling, and they may also be able to form heteromultimers. |
| cd00063 | FN3 | 6.49e-09 | 561 | 648 | 1 | 91 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB29926.1 | 0.0 | 1 | 898 | 1 | 899 |
| QIM09902.1 | 3.06e-213 | 54 | 894 | 28 | 869 |
| QUT88791.1 | 3.00e-197 | 5 | 858 | 15 | 852 |
| ALJ60208.1 | 5.21e-195 | 5 | 858 | 15 | 852 |
| ASB47762.1 | 7.82e-189 | 44 | 851 | 51 | 837 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QVS_A | 7.37e-14 | 699 | 859 | 55 | 227 | ChainA, S-layer domain-containing protein [Acetivibrio thermocellus ATCC 27405] |
| 3MPC_A | 9.79e-07 | 559 | 650 | 3 | 94 | ChainA, Fn3-like protein [Acetivibrio thermocellus ATCC 27405],3MPC_B Chain B, Fn3-like protein [Acetivibrio thermocellus ATCC 27405] |
| 5G56_A | 1.14e-06 | 545 | 669 | 607 | 729 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 4.85e-09 | 550 | 650 | 920 | 1017 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P16950 | 2.39e-06 | 559 | 650 | 927 | 1016 | Amylopullulanase OS=Thermoanaerobacter thermohydrosulfuricus OX=1516 GN=apu PE=1 SV=1 |
| P38939 | 5.39e-06 | 559 | 650 | 926 | 1015 | Amylopullulanase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=apu PE=1 SV=2 |
| A0P8X0 | 9.04e-06 | 549 | 746 | 802 | 979 | Alpha-amylase OS=Niallia circulans OX=1397 GN=igtZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
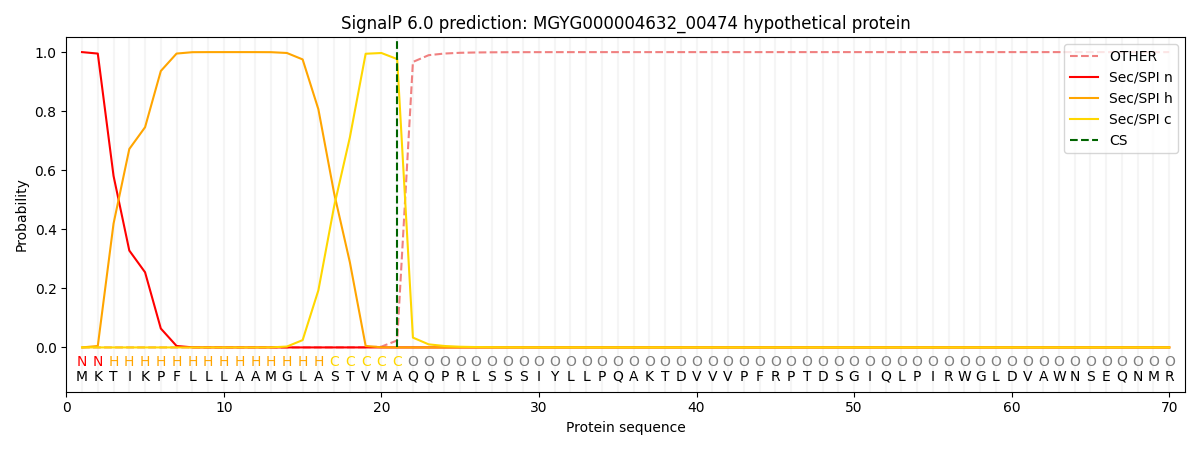
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000392 | 0.998779 | 0.000249 | 0.000198 | 0.000184 | 0.000173 |
