You are browsing environment: HUMAN GUT
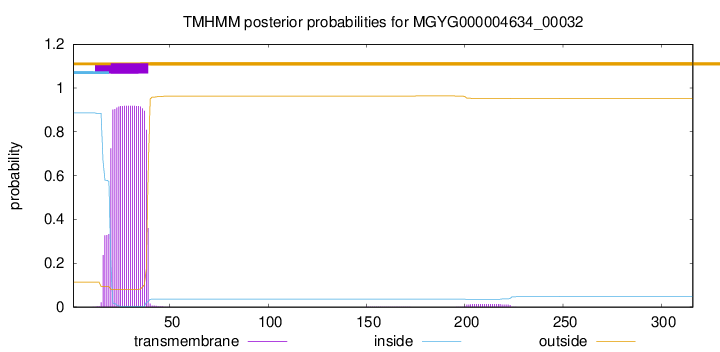
MGYG000004634_00032
Basic Information
help
Species
Lineage
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441;
CAZyme ID
MGYG000004634_00032
CAZy Family
GT32
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004634
2025534
MAG
Germany
Europe
Gene Location
Start: 3757;
End: 4707
Strand: +
No EC number prediction in MGYG000004634_00032.
Family
Start
End
Evalue
family coverage
GT32
98
172
4.4e-22
0.8777777777777778
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam05704
Caps_synth
2.14e-92
38
309
5
276
Capsular polysaccharide synthesis protein. This family consists of several capsular polysaccharide proteins. Capsular polysaccharide (CPS) is a major virulence factor in Streptococcus pneumoniae.
more
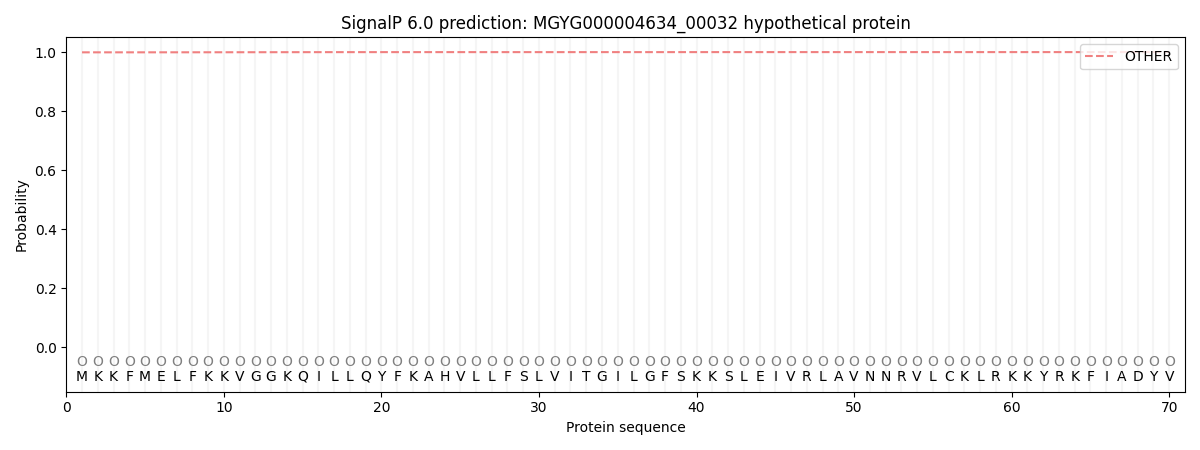
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999316
0.000549
0.000006
0.000001
0.000001
0.000149