You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004636_00640
You are here: Home > Sequence: MGYG000004636_00640
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paramuribaculum sp009775605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum; Paramuribaculum sp009775605 | |||||||||||
| CAZyme ID | MGYG000004636_00640 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36616; End: 38754 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 6.51e-20 | 7 | 316 | 180 | 481 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 7.00e-10 | 10 | 108 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 0.001 | 3 | 231 | 203 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO25184.1 | 3.09e-312 | 3 | 712 | 185 | 924 |
| QKH84924.1 | 1.70e-301 | 3 | 712 | 198 | 923 |
| QRM69893.1 | 1.70e-301 | 3 | 712 | 198 | 923 |
| AUI45420.1 | 2.41e-301 | 3 | 712 | 198 | 923 |
| QTO27419.1 | 2.41e-301 | 3 | 712 | 198 | 923 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D1P_A | 1.66e-10 | 38 | 253 | 240 | 455 | Apostructure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii],6D1P_B Apo structure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii] |
| 6BYC_A | 4.80e-07 | 10 | 231 | 215 | 449 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYE_A | 4.80e-07 | 10 | 231 | 215 | 449 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 4.80e-07 | 10 | 231 | 215 | 449 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 8.29e-07 | 10 | 235 | 213 | 457 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
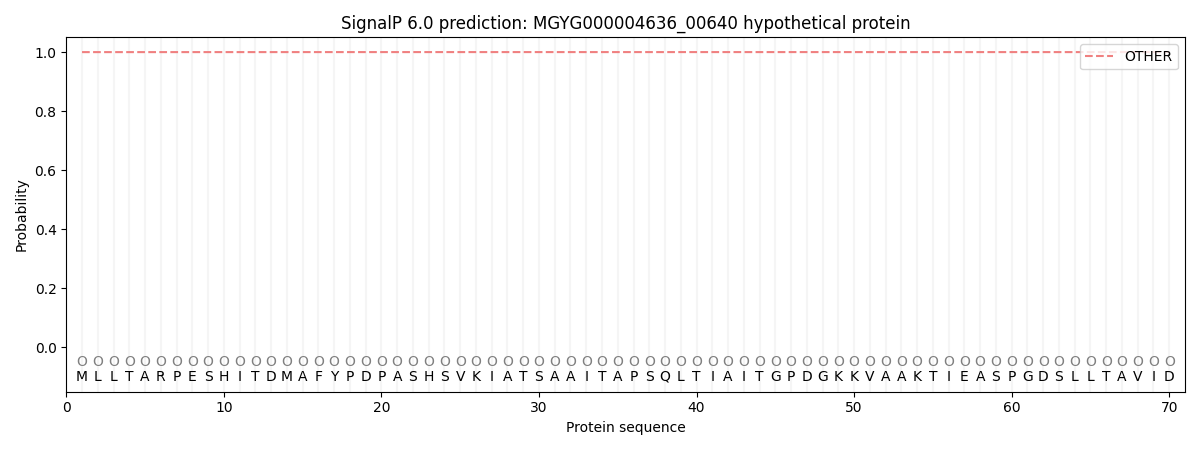
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000028 | 0.000021 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
